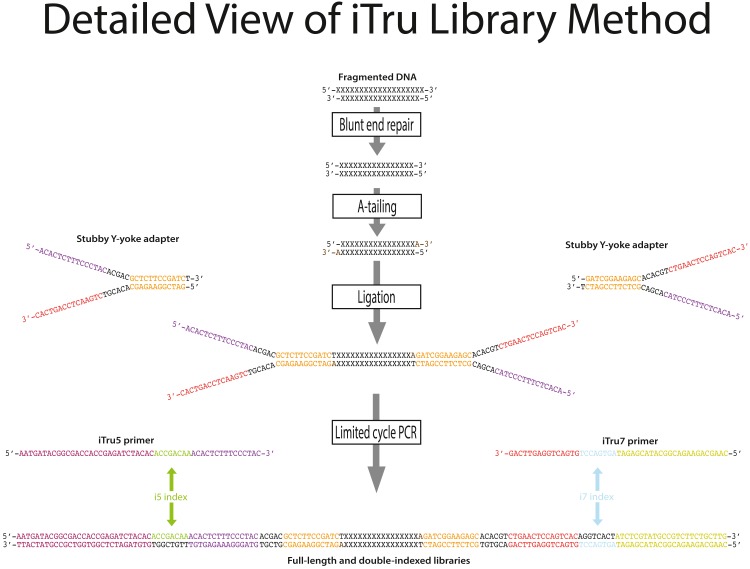
Figure 3. Detailed steps for iTru library construction with relevant sequences.
Starting material is sheared, double-stranded DNA (represented as X) with ragged ends. The DNA is made blunt and 5′ phosphates are added (phosphates not shown). Third, a single adenosine (A) is added to each 3′ end to allow for complementary hybridization of adapters. Next, stubby Y-yoke adapters with complementary ends are ligated to each end of the DNA molecule. These adapters contain both complementary and non-complementary sequences (non-complementary indicated by the gap between the top and bottom strand). These non-complementary sequences include primer-binding sites, as indicated by the colors, used in the next step. In the final step of library preparation, limited-cycle PCR is performed using two distinct primers complementary to the ends of the Y-yoke adapter (shown as iTru5 and iTru7). The primers contain unique indexes (i5 and i7, respectively, shown in color) as well as the P5 and P7 sequences (for color scheme and explanation of functions, see Fig. 1). The index strand in color indicates the sequence of the primer (which is the same as the index read for i5, but the reverse complement is obtained for the i7 index read; see Fig. 4). Note that iNext libraries are similar, except that cytosines are added to the template DNA (instead of adenosines), the Y-yoke adapter has single guanosine overhangs, and the Read1 and Read2 portions have different sequences (cf. Fig. S4).