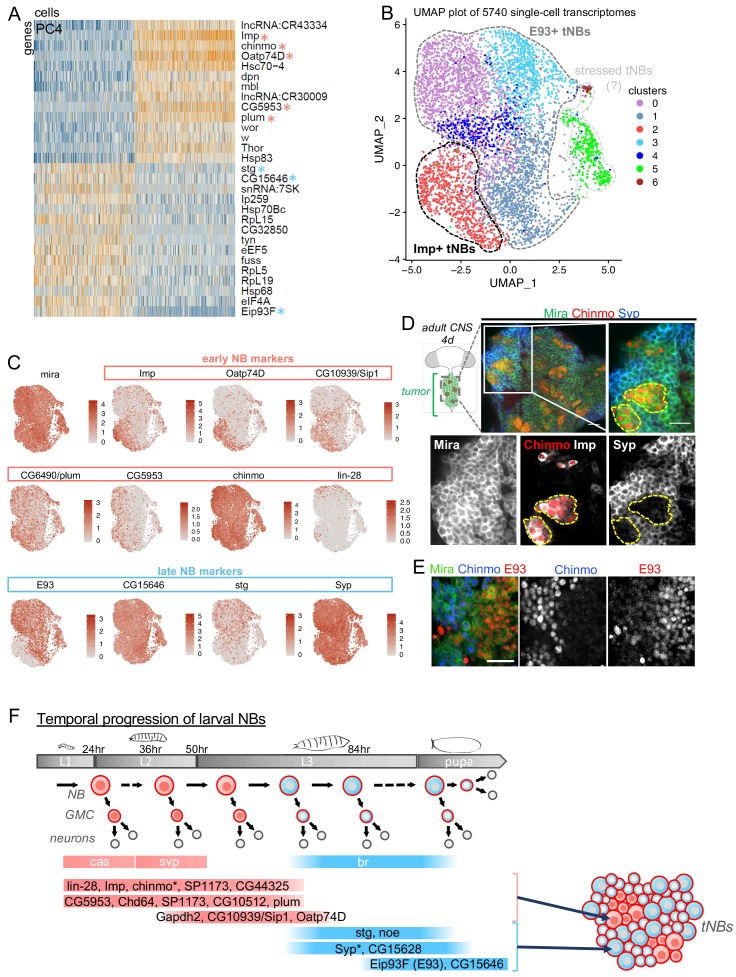
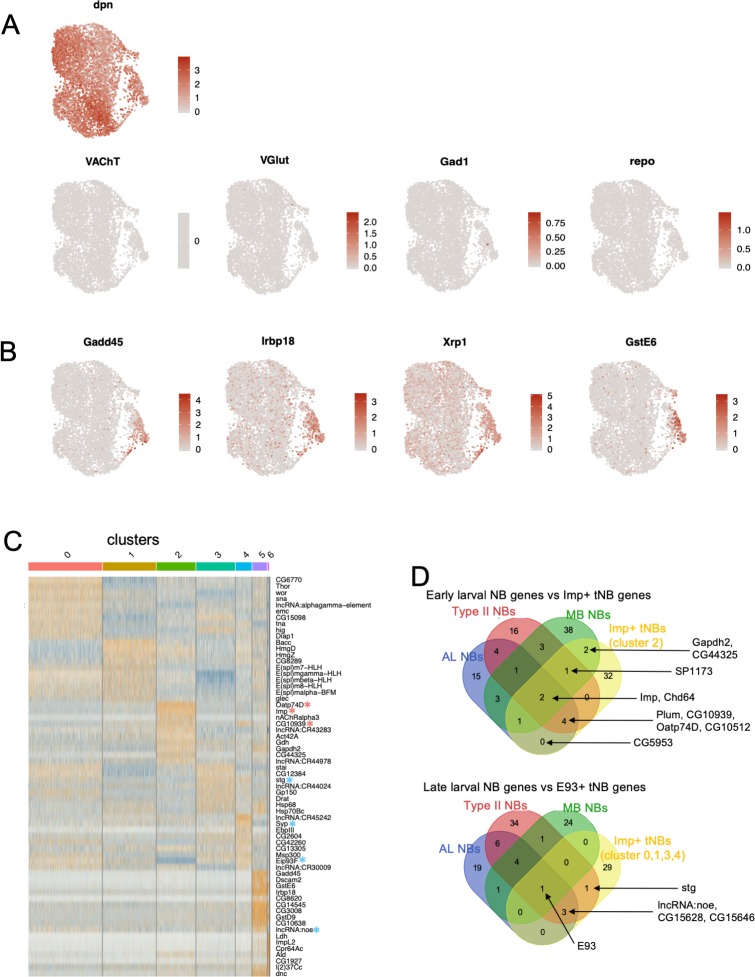
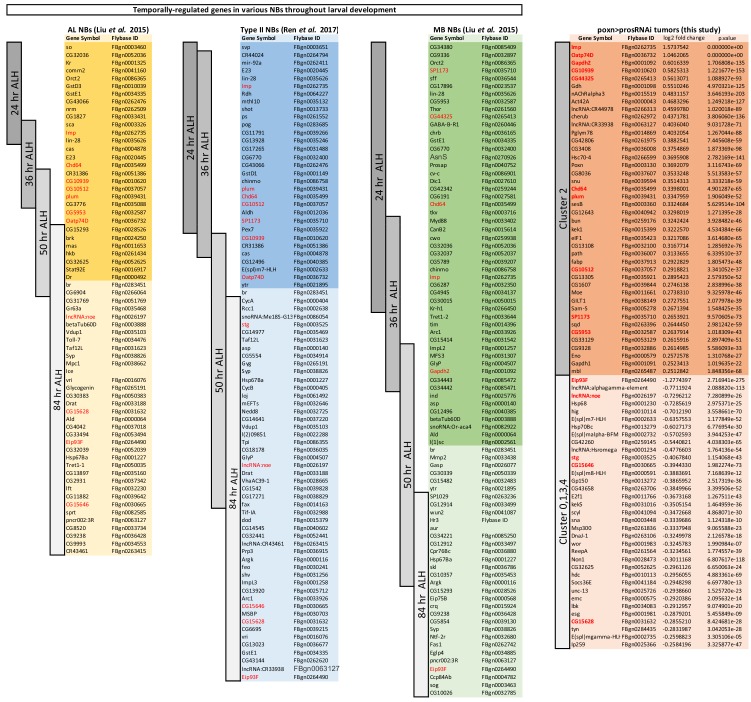
Figure 1. Single-cell RNA-seq identifies a subset of temporal patterning genes that are redeployed in NB tumors defining heterogeneity in NB states.
(A) PCA was performed on single-cell tNB transcriptomes to reduce the dimensions of the data for further analysis. Genes (rows) and cells (columns) are ordered by their PCA scores, and the 500 most extreme cells and 30 most extreme genes on both sides of the distribution are shown in the heatmap. PC4 reveals that tNBs can be discriminated by the expression of early (light red asterisks) vs late (light blue asterisks) larval NB temporal patterning genes. Other larval temporal genes are found in PC3 and PC7 (Figure 1—figure supplement 1C). (B) The UMAP representation of all single cells included in our analysis shows the separation of different clusters. We used a k-nearest neighbor algorithm to call seven clusters, which are shown in different colors on the UMAP plot. (C) Expression of Mira, early and late temporal markers on the UMAP map shown in B. (D) Cartoon representing a ventral view of an adult CNS containing a NB tumor induced during larval stages in the VNC. Green circles are tNBs. Green circles colored in red represent Chinmo+Imp+ tNBs. Immunostainings with anti-Chinmo, anti-Imp and anti-Syp, indicating that Imp/Chinmo and Syp are expressed in a complementary pattern in poxn > prosRNAi tumors found in 4-day-old adults. tNBs are labeled with Mira. Scale bar 20 µm. (E) Immunostainings with anti-Chinmo and anti-E93 indicating that these two transcription factors are expressed in a complementary pattern in poxn > prosRNAi tumors found in 4-day-old adults. tNBs are marked with anti-Mira. Scale bar 20 µm. (F) The light red and blue colors respectively designate ‘early’ and ‘late’ larval NB genes as determined by Liu et al. (2015), Syed et al. (2017) and Ren et al. (2017). A subset of these genes are redeployed in tumors to define distinct tNB states. Asterisks mark temporal patterning genes regulated at the post-transcriptional level in tNBs. Note that cas and svp transcription is associated with early Imp+ NBs, while br transcription is associated with late E93+ NBs during larval stages. In contrast, cas, svp and br transcription do not distinguish Imp+ and E93+ tNBs.