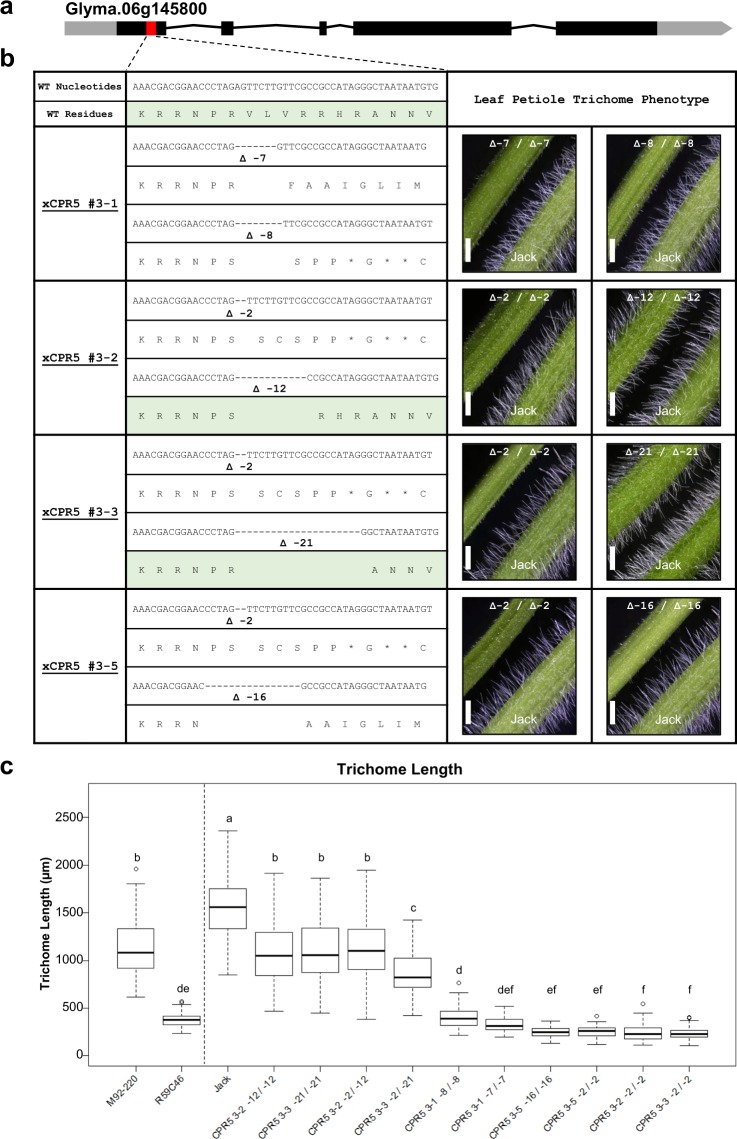
Figure 4.
CRISPR/Cas9 mutation of GmCPR5 (gene model Glyma.06g145800) and phenotypic validation of the candidate gene. (a) A gene model of GmCPR5 with the CRISPR/Cas9 target site indicated with a red box. (b) Nucleotide and predicted translation sequences of GmCPR5 at the CRISPR target site in a wild-type (WT) ‘Jack’ line and in four independently derived regenerated transgenic T2 families in the ‘Jack’ background, named xCPR5 #3-1, xCPR5 #3-2, xCPR5 #3-3, and xCPR5 #3-5. Each of the T0 plants, that gave rise to each T2 family, was found to have biallelic mutations when tested at 30 weeks after transformation. Some mutations were identical among the different plants, resulting in a total of six unique mutated alleles. Two alleles were in-frame deletions, indicated by green shading. All other alleles caused frameshifts. To the right of the sequences are leaf petiole trichome phenotypes of homozygous CRISPR-mutated T2 plants, compared to the ‘Jack’ control. Both alleles in the families xCPR5 #3-1 and xCPR5 #3-5 as well as the 2 bp deletion allele in families xCPR5 #3-2 and xCPR5 #3-3 were frameshift mutations and resulted in very short trichomes, indicating that complete disruption of GmCPR5 results in shorter trichomes. In contrast, the in-frame deletions alleles of the families xCPR5 #3-2 (−12 bp) and xCPR5 #3-3 (−21 bp) resulted in phenotypes more similar to wild-type. The size bar in each image is 2 mm. (c) Trichome length measurements for the lines in the study: wild-type line M92-220, fast neutron mutant R59C46, wild-type line ‘Jack’, and ‘Jack’ derived CRISPR/Cas9 mutant lines. Lines to the left of the vertical dotted line are derived from M92-220 and lines to the right of the line are derived from the cultivar ‘Jack’. The frame-shift CRISPR/Cas9 mutant lines displayed trichome lengths similar to the fast neutron mutant R59C46 validating the gene’s function in trichome length. Uniquely, two homozygous in-frame deletions (−12/−12) and (−21/−21) displayed heritable intermediate trichome length phenotypes. These lines demonstrate that CRISR/Cas9 can be used to create in-frame deletions that result in intermediate phenotypes.