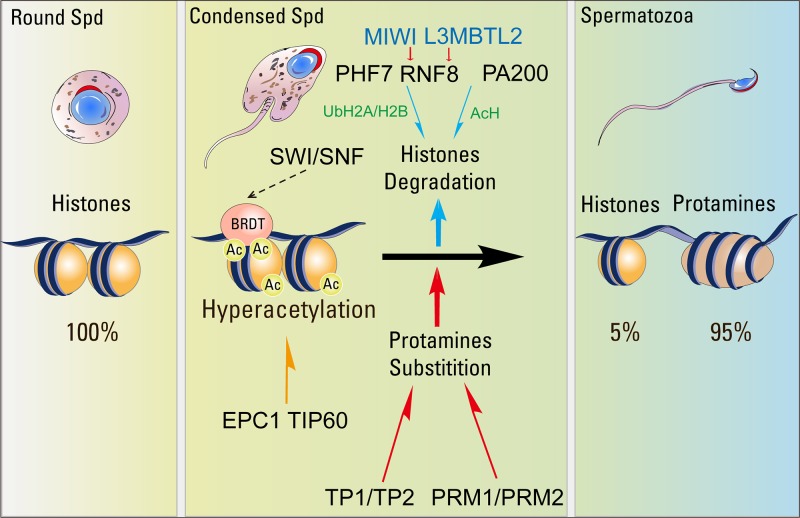
Figure 2.
The key factors related to the histone-to-protamine transition. Global incorporation of various H2A, H2B and H3 histone variants creates highly unstable nucleosomes, which then undergo histone hyperacetylation by EPC1/TIP60 or some other nucleosome acetyltransferase complexes. Acetylation at critical lysines further destabilizes the nucleosomes, while tail acetylation generates a platform for the recruitment of BRDT. BRDT interacts with the SWI/SNF family protein then starts the process of histone eviction and replacement by TPs. Evicted acetylated histones would then be recognized by PA200 and degraded by proteasomes during spermatogenesis. RNF8 could catalyze the ubiquitination of H2A. Ubiquitinated H2A and H2B control H4K16ac by regulating the association of MOF to the chromatin and facilitates histone removal in elongating spermatids. MIWI binds to RNF8 in the cytoplasm of early spermatids (Spd) through a Piwi-interacting RNA (piRNA)-independent manner, and promotes the nuclear translocation of RNF8 in late spermatids to catalyze histone ubiquitination and trigger histone removal. L3MBTL2 could interact with RNF8 and facilitate RNF8-dependent histone ubiquitination-related histone removal. PHF7 could recognize the H3K4me3/me2 and catalyze H2A ubiquitination to facilitate histone removal in elongating spermatids.