Abstract
Leucine-rich repeat kinase 2 (LRRK2) is a promising therapeutic target for the treatment of Parkinson's disease (PD), and orally bioavailable, brain penetrant and highly potent LRRK2 kinase inhibitors are in early stages of clinical testing. Detection of LRRK2 phosphorylation, as well as phosphorylation of Rab10, a LRRK2 kinase substrate, have been proposed as target engagement biomarkers for LRRK2 inhibitor clinical trials. However, these readouts do not seem able to stratify patients based on enhanced LRRK2 kinase activity. Here, we describe a robust cell biological assay based on centrosomal cohesion alterations which were observed in peripheral blood mononuclear cell-derived lymphoblastoid cell lines (LCLs) from patients with G2019S LRRK2 mutations as compared with healthy controls, and could also be detected in a subset of sporadic PD patient samples. We suggest that LCLs may be a valuable resource for LRRK2 research, and that determination of centrosomal cohesion deficits may assist in the stratification of a subset of sporadic PD patients.
Keywords: centrosome cohesion, leucine-rich repeat kinase, protein phosphorylation, Rab protein
Introduction
Parkinson's disease (PD) is a neurodegenerative disorder which affects ∼1% of people over 65 years [1]. Apart from ageing, environmental as well as genetic factors contribute to PD risk. Mutations in several genes have been shown to cause monogenic forms of PD accounting for 5–10% of all PD cases, whilst additional common variants are associated with increased PD risk [2–4]. Amongst monogenic forms, autosomal-dominant point mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are the most frequent, and are also found in a subset of sporadic PD cases [5–7]. Additionally, common protein-coding and non-protein-coding variants at the LRRK2 locus comprise one of the major genetic susceptibility factors for PD, implicating LRRK2 as an important player in the pathogenesis of both familial and sporadic PD [8,9].
LRRK2 is a large protein with multiple domains including a catalytic GTPase and kinase domain [10]. The most common pathogenic missense mutation (G2019S) within the kinase domain has been consistently shown to display increased kinase activity [11]. Similarly, all other currently described pathogenic mutations also seem to increase kinase activity in vivo [12], suggesting that targeting this activity may hold promise for possible disease-modifying strategies, at least with respect to LRRK2-related PD. Towards this goal, highly selective, brain-penetrant and orally bioavailable LRRK2 kinase inhibitors have been synthesized, and some of them are in the early stages of clinical testing [13–15].
Various approaches have been pursued towards establishing assays to monitor the efficacy and target engagement of LRRK2 kinase inhibitors. Initially, and in the absence of validated LRRK2 kinase substrates, studies have focused on analyzing the phosphorylation of LRRK2 itself. Many protein kinases regulate their activity via autophosphorylation [16]. LRRK2 autophosphorylation occurs on Ser1292 which seems to correlate well with kinase activity [17]. However, currently available phospho-specific antibodies are unable to reliably detect this autophosphorylation in an endogenous context [17]. LRRK2 has also been shown to be constitutively phosphorylated by other kinases at a cluster of N-terminal residues, most prominently Ser935 [18,19]. Dephosphorylation of Ser935 has been consistently observed in the presence of a variety of LRRK2 kinase inhibitors, but the phosphorylation state of this site does not change, or even decreases, in the context of various pathogenic LRRK2 mutations [18,20–23]. Thus, whilst being a reliable pharmacodynamic marker to assess the in vivo efficacy of LRRK2 kinase inhibitors in cells and animal models [24–26], LRRK2 Ser935 phosphorylation does not correlate with the intrinsic cellular kinase activity of LRRK2, calling for a better readout for such intrinsic activity.
Recent studies have identified validated physiological substrates for the LRRK2 kinase activity, namely a subset of Rab GTPases including Rab3, Rab8, Rab10, Rab12, Rab35 and Rab43 [12,27–29]. One of the most robust LRRK2 kinase substrates is Rab10, and it is phosphorylated on Thr73 in the switch II region which is important for regulating Rab10 protein interactions [12]. A highly specific and exquisitely sensitive antibody against phosphorylated Rab10 suitable for Western blotting has been recently developed [30]. Both LRRK2 and Rab10 have been shown to be expressed in peripheral blood cells including B-lymphocytes, monocytes and neutrophils, and Rab10 phosphorylation is decreased in these cells upon ex vivo LRRK2 kinase inhibitor treatment [28,31]. Thus, detection of phospho-Rab10 from human peripheral blood-derived cells may allow for improved monitoring of the pharmacokinetics and target engagement of LRRK2 kinase inhibitors in clinical trials [31], even though recent studies have questioned this notion [32].
Ideally, alterations in phospho-Rab10 levels are expected to track with the increase in LRRK2 kinase activity underlying LRRK2-related PD pathogenesis. Determination of the effect size of LRRK2 kinase activity in mouse models homozygous for the G2019S LRRK2 mutation suggest a roughly two-fold increase in Rab10 phosphorylation, and thus an expected 1.5-fold increase in Rab10 phosphorylation in the heterozygous state of G2019S LRRK2 patient-derived samples [12,30]. However, no consistent alterations in the levels of phospho-Rab10 have been detected in neutrophils from G2019S LRRK2-PD patients as compared with healthy controls, possibly confounded by the observed large biological variations amongst the distinct patient-derived samples [31].
We reasoned that relatively small changes in overall LRRK2-mediated Rab phosphorylation may display profound effect sizes in cell biological readouts, especially if due to toxic, gain-of-function type mechanisms. Interestingly, in their phosphorylated state, both Rab8a and Rab10 have been reported to interact with the primary ciliogenesis regulator RILPL1, leading to deficits in ciliogenesis [33]. RILPL1 is localized to the mother centriole, and has been shown to recruit phosphorylated Rab10 to this centrosomal location [33]. In addition, our recent data indicate that pathogenic LRRK2 causes deficits in the cohesion of duplicated centrosomes in dividing cells, in a manner at least in part dependent on Rab8a phosphorylation, and associated with the pericentrosomal/centrosomal accumulation of phosphorylated Rab8a [34]. Centrosomal cohesion deficits were also observed in a small sample of PBMC-derived lymphoblastoid cell lines (LCLs) from G2019S LRRK2-PD patients as compared with healthy controls [34], indicating that such cellular readout may provide a sensitive means towards detecting aberrant LRRK2-mediated Rab phosphorylation events in peripheral patient-derived cells.
Here, we have explored the possibility of assessing endogenous LRRK2 kinase activity in peripheral blood-derived LCLs by monitoring centrosomal cohesion deficits in a larger sampling from G2019S LRRK2-PD patients, as well as in samples derived from sporadic PD patients without the G2019S LRRK2 mutation as compared with controls. Centrosomal cohesion deficits were observed in all G2019S LRRK2-PD patient LCLs examined, and also in a subset of sporadic PD patients. Cohesion deficits were reverted in all cases by a specific LRRK2 kinase inhibitor, suggesting that they are LRRK2 kinase activity-mediated. In contrast, and similar to what has been previously reported in peripheral blood neutrophils [31] as assessed by Western blotting techniques, no significant changes were observed in phospho-Rab10 or in total LRRK2 levels in LCLs from G2019S LRRK2 or sporadic PD patients as compared with healthy controls. Altogether, our data indicate that the cellular readout of centrosomal cohesion deficits from peripheral blood-derived LCLs may be a sensitive measure of altered LRRK2 kinase activity, and able to identify patients which may also benefit from LRRK2 kinase inhibitor-targeted approaches.
Materials and methods
Study participants
Subjects were recruited at the Movement Disorder Unit of the Lille University Hospital with written informed consent, and the study was approved by the local ethics committee. Patients were examined by neurologists specialized in movement disorders. The Movement Disorders Society Unified Parkinson's Disease Rating Scale (MDS-UPDRS) part III was used to define patient motor symptom severity, and l-dopa-equivalent dose (LED) calculated for all patients. Subjects participating in the study donated blood samples for DNA extraction, routine LRRK2 genotyping [35] and lymphocyte immortalization. Six patients heterozygous for the G2019S LRRK2 mutation, 13 sporadic PD patients and 13 unrelated, gender- and age-matched controls were included, and an additional three gender- and age-matched control LCLs (ND01087, ND02550, ND01757), and six gender- and age-matched LCLs from heterozygous G2019S LRRK2 patients (ND01618, ND00075, ND14317, ND02752, ND03000, ND00264) were obtained from the NINDS Coriell Cell Repository (Table 1). PD family history in the three groups was defined as the presence of at least 1 PD patient in 1–3 generations of the family. None of the control or sporadic PD patients had a family history of PD, and all were found negative for the G2019S mutation. Of the six G2019S LRRK2-PD patients recruited to the study, five had a PD family history, with three patients belonging to the same family, with affected members over three generations. Most of the study participants, and 100% of the sporadic PD patients, were of Caucasian origin.
Table 1. Demographic details for Parkinson's disease and control patients.
| Controls | G2019S Parkinson's disease | Sporadic Parkinson's disease | |
|---|---|---|---|
| Participant number | 16 | 12 | 13 |
| Age (y) | 60 ± 9 (42–76) | 63 ± 10 (46–77) | 64 ± 8 (50–77) |
| Sex (% male) | 50% | 50% | 54% |
| Disease duration (y) | - | 9 ± 5 (1–19) | 9 ± 7 (1–27) |
| Age at diagnosis (y) | - | 59 ± 11 (45–74) | 57 ± 9 (40–72) |
| Disease severity | - | 15 ± 5 (5–19) | 18 ± 10 (3–35) |
| LED | - | 633 ± 547 (0–1370) | 1027 ± 663 (0–2300) |
Data shown are mean ± s.d., with the range indicated in parentheses. Disease severity was measured using the Movement Disorders Society Unified Parkinson's Disease Rating Scale (MDS-UPDRS) part III, as well as Hoehn & Yahr (not shown). LED is the calculated L-dopa-equivalent dose. For the six G2019S LRRK2 LCLs from Coriell Cell Repository, information regarding age at diagnosis, UPDRS part III, Hoehn & Yahr and LED was not available.
Peripheral blood mononuclear cell (PBMC) isolation and transformation
Blood samples were collected in BD Vacutainer CPT Cell Preparation Tubes containing sodium heparin (Le Pont-de-Claix, France). Peripheral blood mononuclear cells (PBMCs) were collected and processed according to the manufacturer's instructions, and lymphocytes were immortalized by infection with Epstein–Barr virus (EBV) as described [36]. Briefly, cell lines were established from freshly isolated or cryopreserved lymphocytes using standard EBV transformation protocols which include cell separation by gradient centrifugation and lymphocyte growth enhancement by 1% (v/v) of the mitogenic phytohemagglutinin-M (PHA-M, ThermoFisher Scientific, 10576015). Cell lines were maintained in RPMI 1640 medium supplemented with 20% fetal bovine serum, 2% l-glutamine, 20 units/ml penicillin and 20 µg/ml streptomycin in T75 flasks in 5% CO2 at 37°C. Cells were maintained at a density of 106 cells/ml, with cell density monitored every other day using trypan blue staining.
Immunofluorescence and laser confocal imaging
Coverslips (13 mm diameter) were placed into 24-well plates and coated with Cell-Tak Cell and Tissue Adhesive solution (Corning, nr. 354240) according to manufacturer's instructions. After 20 min incubation at room temperature, the solution was removed, and coverslips were rinsed twice with distilled water followed by air-drying. Lymphoblast cells (200,000/well) were added to dry coated coverslips, and cells attached by slight centrifugation at 690 g for 10 min at room temperature (without brake). Cells were fixed with 2% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 20 min at room temperature, followed by 5 min of ice-cold methanol fixation before permeabilization with 0.2% Triton-X100/PBS for 20 min. After blocking for 1 h in 0.5% BSA (w/v) in 0.2% Triton-X100/PBS (blocking buffer), cells were incubated with primary antibodies overnight at 4°C in blocking buffer. Primary antibodies included mouse monoclonal anti-γ-tubulin (1 : 1000; Abcam ab11316), rabbit polyclonal anti-pericentrin (1 : 1000; Abcam ab4448) or rabbit monoclonal anti-phospho-Rab10 (1 : 100; Abcam ab237703). The following day, coverslips were washed three times 10 min in 0.2% Triton-X100/PBS, followed by incubation with secondary antibodies (Alexa488-conjugated goat anti-mouse (1 : 1000; Invitrogen) and Alexa594-conjugated goat anti-rabbit (1 : 1000; Invitrogen)) in blocking buffer for 1 h. Coverslips were washed three times in 0.2% Triton-X100/PBS, rinsed in PBS and mounted in mounting medium with DAPI (Vector Laboratories). For staining with the anti-phospho-Rab10 antibody, the methanol fixation was omitted, and 0.2% Triton-X100 was replaced by 0.1% saponin throughout. For treatment of cells with MLi2, cells were seeded on coated coverslips as described above, and incubated with or without MLi2 (generous gift of D. Alessi, University of Dundee, U.K.) (or DMSO as control) at the indicated concentrations for two hours at 37°C, before attachment by slight centrifugation followed by fixation as described above.
Images were acquired on a Leica TCS-SP5 confocal microscope using a 63× 1.4 NA oil UV objective (HCX PLAPO CS). Images were collected using single excitation for each wavelength separately, and dependent on secondary antibodies (488 nm Argon Laser line and a 498–551 nm emission band pass; 543 HeNe laser line and a 615–671 nm emission band pass; 405 nm UV diode and a 418–467 nm emission band pass). Around 30 image sections of selected areas were acquired with a step size of 0.5 µm, and z-stack images analyzed and processed using Leica Applied Systems (LAS AF6000) image acquisition software. The same laser intensity settings, exposure times and zoom were used for image acquisition of individual experiments to be quantified.
The mean distance between duplicated centrosomes in control cells was 1.3 ± 0.2 µm (mean ± s.e.m., n = 10 cells), and duplicated centrosomes were scored as being separated when the distance was >1.3 µm [34]. Only cells which displayed clear centrosomal staining by both the anti-pericentrin and anti-γ-tubulin antibodies were analyzed, and in all cases, mitotic cells were excluded from this analysis. For each sample, minimally 100 cells were quantified by an observer blind to condition. Additionally, some experimental conditions were independently quantified by an additional two observers blind to condition, with identical results obtained in all cases.
Quantification of the phospho-Rab10 signal was performed over non-processed and non-saturated images acquired during the same day with the same laser intensities. For intensity analyzes, integrated densities of 100 cells per line per experiment were determined. Values were background-corrected, and normalized to the area of the 3 µm ROI which was placed over the centrosome as identified by pericentrin staining. The analysis was performed by Fiji.
Cell extracts and Western blotting
One million cells were centrifuged at 1030 g for 5 min at 20°C, the cell pellet was washed once with HBSS (Hanks Buffered Salt Solution; 140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0.4 mM MgSO4-7H2O, 0.5 mM MgCl2-6H2O, 0.3 mM Na2HPO4-2H2O, 0.4 mM KH2PO4, 6 mM glucose, 4 mM NaHCO3, pH 7.2) and followed by resuspension in 100 µl of lysis buffer (1% Triton-X100, 150 mM NaCl, 20 mM TRIS-EDTA pH 7.4, 1 mM EDTA, 10% glycerol, phosphatase inhibitor cocktail (Sigma, P5726) and protease inhibitor cocktail (Roche, nr. 046931160001)). Extracts were incubated during 30 min at 4°C on a rotary wheel, followed by a short three-pulse sonication. A small sample was taken for determination of protein concentration, and the remainder of extract snap-frozen in liquid N2 and stored at −80°C. Protein concentration was estimated using BCA assay (Pierce) according to manufacturer's specifications. For treatments with MLi2, one million cells were incubated with or without 10 nM MLi2 (or DMSO as control) for 2 h at 37°C, followed by centrifugation and processing as described above.
LCL extracts were mixed with Nu PAGE LDS sample buffer (Novex, Life Technologies, NP0008) supplemented with β-mercaptoethanol to a final volume of 2.5% (v/v), and heated at 70°C for 10 min. Twenty micrograms of samples were loaded onto 4–20% precast polyacrylamide gels (Bio-Rad, 456–1096) and electrophoresed at 100 V for 2 h with SDS running buffer (Tris-Glycine Running Buffer; 25 mM TRIS pH 8.6, 190 mM glycine, 1% SDS). At the end of electrophoresis, proteins were electrophoretically transferred onto PVDF membranes (GE Healthcare) at 40 mA overnight at 4°C in transfer buffer (20 mM TRIS pH 8.6, 122 mM glycine, 20% MeOH (v/v)). The membrane was blocked in blocking buffer (Li-COR Biosciences, Li-COR Odyssey PBS blocking buffer, 927–40000) for 1 h at room temperature, followed by cropping the membrane into three pieces for Li-COR multiplexing (top piece until 75 kD, middle piece until 37 kD, bottom piece). Membranes were washed three times in 0.1% (v/v) Tween-20/PBS, followed by incubation with primary antibodies in 0.2% (v/v) Tween-20 in blocking buffer overnight at 4°C. The top piece was incubated with a rabbit anti-pS935-LRRK2 UDD2 antibody (1 : 500, Abcam, ab133450) multiplexed with a mouse monoclonal anti-LRRK2 antibody (1 : 1000; UC Davies/NIH NeuroMab, clone N241A/34, 75–235). The middle piece was incubated with a mouse monoclonal anti-α-tubulin antibody (1 : 10 000; Sigma, clone DM1A), and the bottom piece was incubated with a rabbit monoclonal anti-phospho-T73-Rab10 antibody (1 : 1000; Abcam, ab230261) multiplexed with a mouse monoclonal total Rab10 antibody (1 : 100; Nanotools, 0680-100/Rab10-605B11). Membranes were washed three times for 10 min in 0.1% Tween-20/PBS, followed by incubation with secondary antibodies for 1 h at room temperature in 0.2% Tween-20 in blocking buffer. Secondary antibodies included goat anti-rabbit IRDye 800CW and goat anti-mouse IRDye 680CW (1 : 14 000). Membranes were washed with 0.1% Tween-20/PBS for three times 10 min each, followed by a wash with PBS only. Blots were imaged via near-infrared fluorescent detection using Odyssey CLx imaging system, and quantification was done using the instrument's Image Studio software. Sample extracts were analyzed 1–3 times, representative immunoblots are shown in the figures, and all immunoblots are available as Supplementary Figure S1.
Flow cytometry assays
For cell cycle analysis, one million cells were centrifuged at 1030 g for 5 min, followed by resuspension in 100 µl of PBS at 4°C, and cells fixed by the addition of 100 µl of 4% PFA/PBS during 15 min on ice. After fixation, cells were centrifuged at 1030 g for 6 min at 4°C, and the cell pellet resuspended in 340 µl PBS, followed by incubation with RNAse A (Roche; 10 109 169 001) (3 µl of 10 mg/ml stock) and propidium iodide (Sigma–Aldrich; P4864; 1 mg/ml) (12 µl of 1 mg/ml stock) for 20 min at 37°C in the dark. Flow cytometry analysis was performed in a FACScaliburTM (Becton Dickinson), propidium iodide-positive signals analyzed using a 670 nm fluorescence emission filter, and data represented with respect to the amount of DNA present per cell.
Analysis of apoptosis was performed using Alexa Fluor-488 AnnexinV/Dead Cell Apoptosis Kit (Invitrogen, V13241) according to the manufacturer's instructions. Briefly, one million cells were centrifuged at 500 g for 3 min, and the pellet washed with 1 ml of ice-cold PBS. Cells (0.5 × 106) were resuspended in 100 µl of binding buffer and labeled with AlexaFluor 488-AnnexinV and propidium iodide for 15 min on ice. Cells were subsequently analyzed by flow cytometry in a FACScaliburTM (Becton Dickinson) using a 488 nm excitation laser and emission at 530 nm (annexinV) and 670 nm (propidium iodide).
Proximity ligation assays
Proximity ligation assays (PLAs) were performed essentially as described [37] using DuoLink® PLA Technology according to manufacturer's instructions (Sigma–Aldrich; Duolink® In Situ PLA probe anti-rabbit PLUS (DUO92002), Duolink® In Situ PLA probe anti-mouse MINUS (DUO92004), Duolink® In Situ Detection Reagents Red (DUO92008)). Briefly, cells on Cell-Tak-coated coverslips were fixed in 4% PFA/PBS as described above, followed by three washes in PBS. Coverslips were blocked in blocking solution followed by incubation with primary antibodies overnight at 4°C. Primary antibodies were rabbit polyclonal anti-phospho-S1292-LRRK2 (1 : 1000; Abcam, ab203181) and mouse monoclonal anti-LRRK2 antibody (1 : 1000; UC Davies/NIH Neuromab, clone 241A/34, 75–235). The proximity ligation signal was visible as individual dots, and analyzed by confocal microscopy as described above. The number of PLA positive dots/cell was quantified from around 300 cells per condition from maximal intensity projections using Leica Applied Systems (LAS AF6000) image acquisition software, and various control experiments included in each assay run.
Statistical analysis
Data were checked for normal distribution using the Shapiro–Wilk test. One-way ANOVA with Tukey's post-hoc test was employed, with significance set at P < 0.05. Non-normally distributed data were analyzed by Kruskal–Wallis test and Dunn's multiple comparison. Spearman correlations were used to determine associations between levels of LRRK2 or LRRK2 Ser935 and Rab10 Thr73. Paired t-test analysis was performed for comparison of the cohesion phenotypes in the presence and absence of MLi2. All statistical analysis and graphs employed the use of Prism software version 7.0 (GraphPad, San Diego, CA).
Results
Centrosomal cohesion deficits in LCLs from G2019S patients do not cause cell cycle alterations
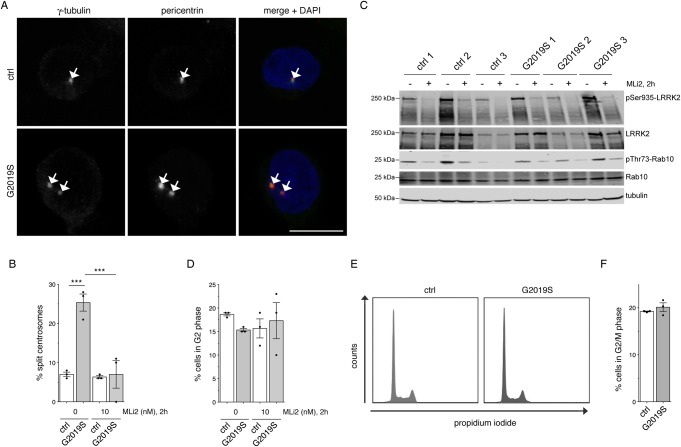
To determine whether centrosomal cohesion deficits may comprise a valid cellular biomarker in peripheral patient-derived cells, we initially used a small set of LCLs from healthy control and G2019S LRRK2-PD patients, and determined centrosomal cohesion deficits from cells stained with an improved protocol employing antibodies against two distinct centrosomal markers (pericentrin and γ-tubulin) (Figure 1A). When quantifying the cells with duplicated centrosomes, all three G2019S LRRK2 LCLs showed a significant increase in the percentage of cells displaying a split centrosome phenotype as compared with control cells, which was reverted upon short-term incubation with the LRRK2 kinase inhibitor MLi2 (Figure 1B). In parallel, we analyzed the effects of MLi2 on LRRK2 phosphorylation as well as on LRRK2 substrate phosphorylation. In all cases, MLi2 treatment significantly reduced both LRRK2 Ser935 phosphorylation as well as Rab10 Thr73 phosphorylation, confirming the on-target effect of the LRRK2 kinase inhibitor (Figure 1C).
Figure 1. Monitoring centrosomal cohesion deficits and cell cycle alterations in a subset of healthy control and G2019S LRRK2-PD LCLs.
(A) Example of healthy control (ctrl) or G2019S LRRK2 PD-derived LCLs (G2019S) stained for two centrosomal markers (γ-tubulin and pericentrin) and DAPI. Scale bar, 10 µm. (B) The centrosome phenotype was quantified from 100 cells per line, and from three control and three G2019S LRRK2 LCL lines. Control or G2019S LRRK2 LCLs were treated with MLi2 (10 nM) for 2 h as indicated. Bars represent mean ± s.e.m.; *** P < 0.005 (one-way ANOVA with Tukey's post-hoc test). (C) Cells were incubated either in the presence or absence of 10 nM MLi2 for 2 h as indicated, and extracts analyzed for LRRK2 Ser935, LRRK2, Rab10 Thr73, Rab10, or tubulin as loading control. Membranes were developed using the Odyssey CLx scan Western blot imaging system, and antibodies multiplexed as described in Materials and methods. (D) Quantification of the percentage of cells displaying duplicated centrosomes, a phenotype mainly reflecting cells in G2 phase (% cells in G2 phase). A total of 100 cells per line were quantified from the three control and three G2019S LRRK2 LCL lines in the absence or presence of MLi2 (10 nM) for 2 h as indicated. (E) Example of flow cytometry traces of one control and one G2019S line upon propidium iodide staining as indicated. (F) Quantification of the percentage of cells displaying duplicated DNA content as assessed by propidium iodide staining (% cells in G2/M phase) from three control and three G2019S LRRK2 LCL lines.
Around 15–20% of all cells displayed duplicated centrosomes, a phenotype which mainly reflects cells in G2 phase of the cell cycle [38], and which was not found to be drastically different between the distinct cell lines (Figure 1D). As an alternative means, we performed cell cycle analysis by flow cytometry, which also did not reveal significant changes between the control and G2019S LRRK2 LCLs, with a similar percentage of cells identified in G2/M phase as when scoring the percentage of cells with duplicated centrosomes by immunocytochemistry (Figure 1E,F). Finally, G2019S LRRK2 LCLs also did not show alterations in apoptosis as compared with control LCLs (Supplementary Figure S2A,B). Therefore, the increased distance between duplicated centrosomes in G2019S LRRK2 LCLs does not impact on cell cycle progression and/or cell viability.
Centrosomal cohesion deficits are reverted by LRRK2 kinase inhibitor in a dose-dependent and dynamic manner
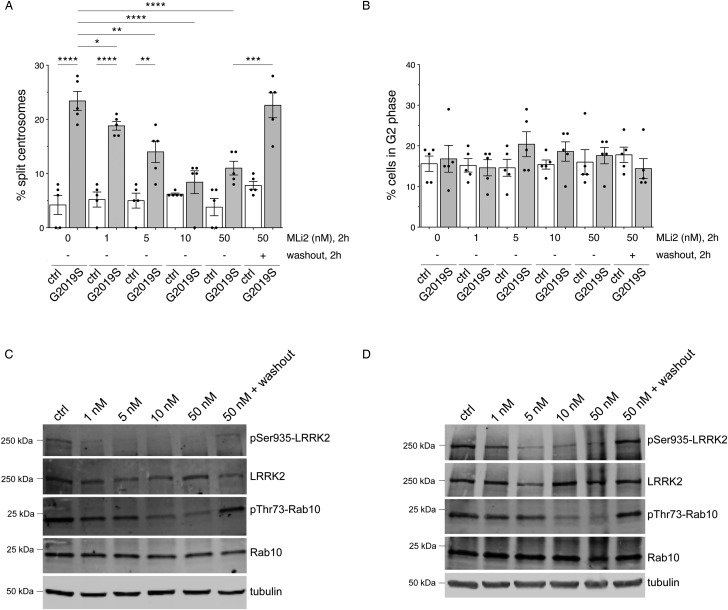
To assess LRRK2 inhibitor dose response, we next treated five distinct control and G2019S LRRK2 LCLs with increasing concentrations of MLi2 for 2 h. None of the concentrations tested altered centrosome cohesion in control cells (Figure 2A). In contrast, the cohesion deficit observed in the G2019S LRRK2 LCLs was progressively reduced at 1 nM and 5 nM, and abolished at 10 nM and 50 nM inhibitor concentrations, respectively (Figure 2A). When incubating cells with 50 nM MLi2 for 2 h, followed by washout into media without inhibitor for another two hours, the centrosomal cohesion deficit in the G2019S LRRK2 LCLs was fully reestablished, indicating the rapid and dynamic nature of this cellular readout (Figure 2A). None of the treatments caused cell cycle alterations as analyzed by scoring the percentage of cells with duplicated centrosomes (Figure 2B). The dose-dependency was well matched with alterations observed in LRRK2 Ser935 phosphorylation and Rab10 Thr73 phosphorylation, with a partial decrease at 1 nM and 5 nM MLi2, a potent decrease at 10 nM and 50 nM MLi2, and a complete reversal upon washout (Figure 2C,D). Such dose-dependent inhibition and reversal of LRRK2 Ser935 phosphorylation and Rab10 Thr73 phosphorylation was observed in both control (Figure 2C) and G2019S LRRK2 LCLs (Figure 2D), even though MLi2 treatment had no effect on centrosome cohesion in the control LCLs (Figure 2A). Thus, LRRK2 kinase inhibitor causes a potent, dose-dependent and reversible inhibition of the centrosomal cohesion deficits in LRRK2 G2019S LCLs, associated with changes in LRRK2 Ser935 phosphorylation and Rab10 Thr73 phosphorylation indicative of on-target inhibition of LRRK2 kinase activity.
Figure 2. Dose response and reversibiltiy analysis of centrosomal cohesion deficits and Rab10 phosphorylation in control and G2019S LRRK2 LCLs.
(A) The centrosome phenotype was quantified from five distinct control and five G2019S LRRK2 LCL lines, in either the presence or absence of the indicated concentrations of MLi2 for 2 h. In addition, cells were treated with 50 nM MLi2 for 2 h, followed by incubation in medium without MLi2 for an additional 2 h (washout) before quantification. Bars represent mean ± s.e.m.; * P < 0.05; ** P < 0.01; *** P < 0.005; **** P < 0.001 (one-way ANOVA with Tukey's post-hoc test). (B) Quantification of the percentage of cells displaying duplicated centrosomes, a phenotype mainly reflecting cells in G2 phase, from a total of 100 cells per line, for each of the five control and five G2019S LRRK2 lines. (C) Control LCL was incubated in either the presence or absence of the indicated concentrations of MLi2 for 2 h as indicated, or treated with 50 nM MLi2 for 2 h followed by incubation in medium without inhibitor for an additional 2 h (washout), and extracts analyzed for LRRK2 Ser935, LRRK2, Rab10 Thr73, Rab10 or tubulin as loading control. Membranes were developed using the Odyssey CLx scan Western blot imaging system, and antibodies multiplexed as described in Materials and methods. (D) Same as (C), but performed with G2019S LRRK2 LCL. Similar results were obtained in two independent experiments.
Analysis of centrosomal cohesion deficits in LCLs from a larger G2019S LRRK2-PD cohort
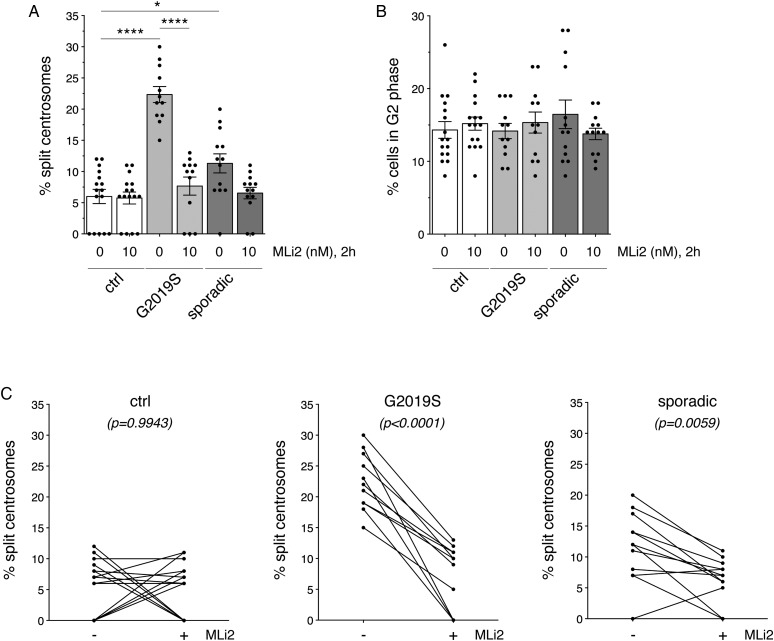
We next sought to probe for centrosomal deficits in a larger sampling of LCLs. A total of 16 age- and sex-matched control group and 12 G2019S LRRK2 LCLs were analyzed, which included an independent determination from the five previously employed control and G2019S LRRK2 LCLs for proper age- and sex-matching purposes (Figure 3A and Table 1). Centrosomal cohesion deficits were observed in all 12 G2019S LRRK2 LCLs as compared with controls (Figure 3A), which was not associated with a change in the percentage of cells displaying duplicated centrosomes (Figure 3B). In all cases, these deficits were reverted upon MLi2 treatment (Figure 3A,C).
Figure 3. Centrosomal cohesion deficits in LCLs from control, G2019S LRRK2-PD and sporadic PD patients.
(A) Centrosomal cohesion deficits were quantified from a total of 16 age- and sex-matched control, 12 G2019S LRRK2-PD and 13 sporadic PD LCLs in either the absence or presence of 10 nM MLi2 for 2 h as indicated. Bars represent mean ± s.e.m.; ** P < 0.01; **** P < 0.001 (one-way ANOVA with Tukey's post-hoc test). (B) Quantification of the percentage of cells displaying duplicated centrosomes from a total of 100 cells per LCL line. (C) Paired t-test analysis of centrosomal cohesion deficits from control, G2019S LRRK2 and sporadic PD LCLs for each cell line in the absence or presence of MLi2 as indicated. Note that differences in the values between 0 and 10% are not significant given the small number of cells displaying a duplicated split centrosome phenotype.
To analyze whether centrosomal cohesion deficits were specific to LRRK2 G2019S as compared with sporadic PD samples, we analyzed LCLs from 13 age- and sex-matched sporadic PD patients negative for the G2019S LRRK2 mutation (Figure 3A,D and Table 1). Interestingly, three out of 13 LCLs from sporadic PD patients displayed a centrosomal cohesion deficit similar to LRRK2 G2019S LCLs, which was reverted upon MLi2 treatment (Figure 3A,C) and was not associated with a change in the percentage of cells displaying duplicated centrosomes (Figure 3B). Therefore, centrosomal cohesion deficits seem to be a robust cellular readout in LCLs from LRRK2 G2019S patients, but also observable in LCLs from a subset of sporadic PD patients, and in all cases dependent on the LRRK2 kinase activity.
Association between centrosomal cohesion deficits, levels of LRRK2 or Rab10 Thr73 phosphorylation, or PD clinical variables
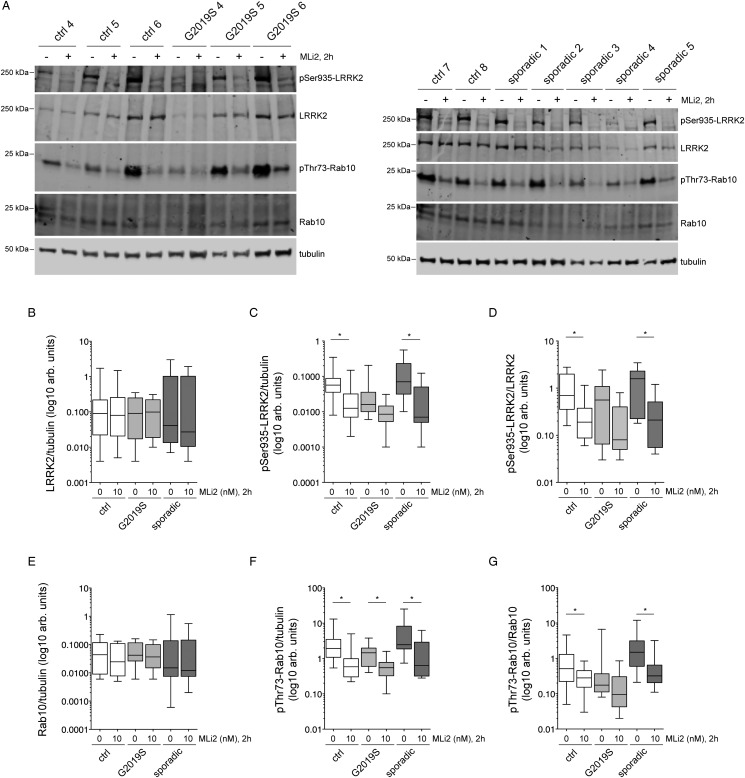
We next wondered whether the centrosomal cohesion deficits observed in G2019S LRRK2 LCLs and sporadic LCLs were associated with increased levels of LRRK2 and/or Rab10 Thr73 phosphorylation, respectively. Levels of total LRRK2 varied drastically amongst individual LCLs, but there were no significant differences in the levels of total LRRK2 between healthy control, LRRK2 G2019S or sporadic PD LCLs (Figure 4A,B), even though some LRRK2 G2019S lines had hardly detectable levels of total LRRK2 (Supplementary Figure S1), reminiscent of the reported decrease in LRRK2 protein levels in postmortem brain extracts from LRRK2-PD patients as compared with control or sporadic PD patients [39]. Levels of LRRK2 Ser935 phosphorylation normalized to either tubulin or to total LRRK2 were similar in LCLs from control, G2019S LRRK2 or sporadic PD patient samples, and were significantly reduced by MLi2 in control and sporadic samples (Figure 4C,D). The total levels of Rab10 protein were generally low, and less variable than LRRK2 protein between individual patient samples, without a difference between genotypes observed (Figure 4E and Supplementary Figure S2). Rab10 Thr73 phosphorylation was not different between control, G2019S or sporadic PD LCLs, but significantly decreased upon 2 h MLi2 treatment in control and sporadic samples (Figure 4F,G). Therefore, LCLs display a large variability in total LRRK2 protein, with more similar levels in total Rab10, but no significant differences in the amount of total proteins and/or phospho-proteins between control, G2019S LRRK2 or sporadic PD samples.
Figure 4. Analysis of LRRK2, LRRK2 Ser935, Rab10 and Rab10 Thr73 phosphorylation in LCLs from control, G2019S LRRK2-PD and sporadic PD patients.
(A) Example of three control and three G2019S LRRK2 LCL lines (left), or two distinct control and five sporadic PD LCL lines (right), treated with or without 10 nM MLi2 for 2 h. Cells were subsequently lysed and extracts subjected to quantitative immunoblot analysis with the indicated antibodies, and membranes developed using Odyssey CLx scan Western Blot imaging system. Note that ‘sporadic 2’ and ‘sporadic 4' are two out of the three sporadic PD LCLs which display a centrosomal cohesion deficit. (B) Control, G2019S LRRK2 and sporadic PD LCL extracts were analyzed as described in (A), and immunoblots quantified for full-length LRRK2/tubulin ratio. (C) Immunoblots of the type depticted in (A) were quantified for LRRK2 Ser935/tublin ratio. * P < 0.05. (D) Immunoblots were quantified for LRRK2 Ser935/LRRK2 ratio. * P < 0.05 (E) Immunoblots were quantified for Rab10/tubulin ratio. (F) Immunoblots were quantified for Rab10 Thr73/tubulin ratio. * P < 0.05. (G) Immunoblots were quantified for Rab10 Thr73/Rab10 ratio. * P < 0.05. Statistical analysis was performed with Kruskal–Wallis test with Dunn's multiple comparison. All data are presented as whisker plots.
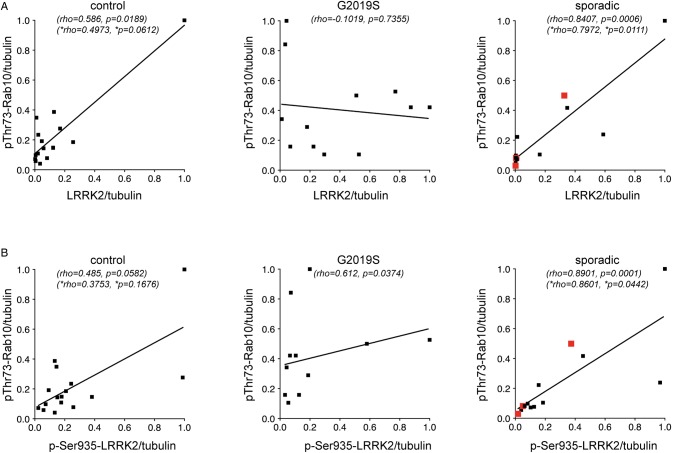
Correlation analyzes were performed to determine possible associations between LRRK2 and phosphorylated Rab10. In control and sporadic PD LCLs, there was a significant correlation between LRRK2 levels and Rab10 Thr73 phosphorylation (Figure 5A). Similarly, in patients with and without the G2019S LRRK2 mutation, there was a significant correlation between LRRK2 Ser935 levels and Rab10 Thr73 phosphorylation (Figure 5B). The lack and/or weak correlation between LRRK2 levels/LRRK2 Ser935 levels and Rab10 Thr73 phosphorylation in the G2019S LRRK2 samples correlated with a tendency of lower total LRRK2 levels in this specific patient sampling analyzed (Figure 5A,B). As correlation coefficients are sensitive to outliers, we recalculated them after taking out the most outlying datapoint, which still yielded a significant correlation between LRRK2 levels or LRRK2 Ser935 levels and Rab10 Thr83 phosphorylation for the sporadic LCL samples, whilst significance was lost in the control samples. Thus, and at least in sporadic PD LCLs, increased LRRK2 levels seem to correlate well with an increase in Rab10 Thr73 phosphorylation levels.
Figure 5. Correlation analysis between levels of LRRK2 or LRRK2 Ser935 and Rab10 Thr73 in LCLs from control, G2019S LRRK2-PD and sporadic PD patients.
(A) Spearman correlation analysis revealed a significant association between LRRK2 levels and Rab10 Thr73 levels in sporadic PD patients. (B) Spearman correlation analysis revealed a significant association between LRRK2 Ser935 levels and Rab10 Thr73 levels in sporadic PD samples. The maximal datapoint value in each genotype was normalized to 1. Rho and P values including all data sets (top), or excluding the outlying datapoint (bottom, *) are indicated for each correlation analysis. Red datapoints indicate the three sporadic PD samples which display a centrosomal cohesion deficit.
Finally, correlation analyzes were performed to determine any associations between LRRK2, Rab10 or their phosphorylation with available PD clinical variables in the sporadic PD samples. No significant correlations were observed between LRRK2 levels, LRRK2 Ser935 levels or Rab10 Thr73 phosphorylation levels and UPDRS motor score, disease duration, age at onset or dopamine medication doses in the sporadic PD patient cohort samples (Supplementary Figure S3). Similarly, there were no correlations between LRRK2 levels, LRRK2 Ser935 levels or Rab10 Thr73 phosphorylation levels and clinical variables in the three sporadic PD patients which displayed a centrosomal cohesion phenotype (Supplementary Figure S3).
Correlation between centrosomal cohesion deficits and LRRK2 activity and/or pericentrosomal/centrosomal phospho-Rab10 accumulation
We wondered whether the centrosomal cohesion deficits observed in G2019S LRRK2 but not control LCLs may correlate with increased LRRK2 kinase activity, rather than with total LRRK2 protein levels. The autophosphorylation site Ser1292 serves as a physiological and direct marker for LRRK2 kinase activity, but current antibodies do not allow detection of this autophosphorylation site by Western blotting at endogenous levels [17]. Therefore, we employed the recently developed PLA to assess Ser1292 phosphorylation of LRRK2, whereby the signal of the anti-Ser1292 antibody is amplified and detected only when in proximity to the signal from a validated total LRRK2 antibody [37].
To independently validate the PLA to detect LRRK2 kinase activity [37], we initially transfected HEK293 cells with GFP-tagged pathogenic Y1699C-LRRK2, which displays robust kinase activity and triggers an activity-mediated centrosomal cohesion phenotype [12,34]. Pathogenic LRRK2 expression caused a significant increase in the phospho-Ser1292 PLA signal intensity, which was abolished upon pretreatment of cells with MLi2 (Supplementary Figure S4A,B). In addition, and as analyzed by quantitative light microscopy, pathogenic LRRK2 expression levels correlated with the PLA signal intensity (Supplementary Figure S4C), suggesting that the signal was a dose-dependent reflection of LRRK2 kinase activity.
We next analyzed the phospho-Ser1292 PLA signal under conditions of endogenous LRRK2 expression levels by using wildtype, mutant LRRK2 knockin (KI) (G2019S/G2019S-KI or R1441G/R1441G-KI), and LRRK2-deficient (LRRK2-KO) HEK293 cells obtained by CRISPR/Cas9 gene editing [37]. As compared with cells transiently overexpressing pathogenic LRRK2, the PLA signal in the distinct HEK293 cell lines expressing endogenous levels of LRRK2 was drastically reduced, and was quantified as the number of individual fluorescence dots per cell, rather than total PLA signal intensity per cell (Supplementary Figure S4D). Such differences in assay signal between HEK293 cells overexpressing mutant LRRK2 as compared with LRRK2-KI cells correlated with differences in LRRK2 protein levels, as well as with LRRK2 kinase activity, as assessed by Western blotting techniques (Supplementary Figure S4E). There was a significant increase in the PLA signal in the LRRK2 R1441G/R1441G-KI cells (Supplementary Figure S4D). In contrast, the assay was unable to detect endogenous LRRK2 kinase activity in the wildtype or G2019S/G2019S-KI cells, as the signal observed was identical with the one obtained in the LRRK2-KO cells, or when omitting one of the two primary antibodies from the PLA reaction (Supplementary Figure S4D). Thus, whilst the assay can serve as a readout for LRRK2 kinase activity, it displays limited sensitivity in the context of endogenous LRRK2 levels.
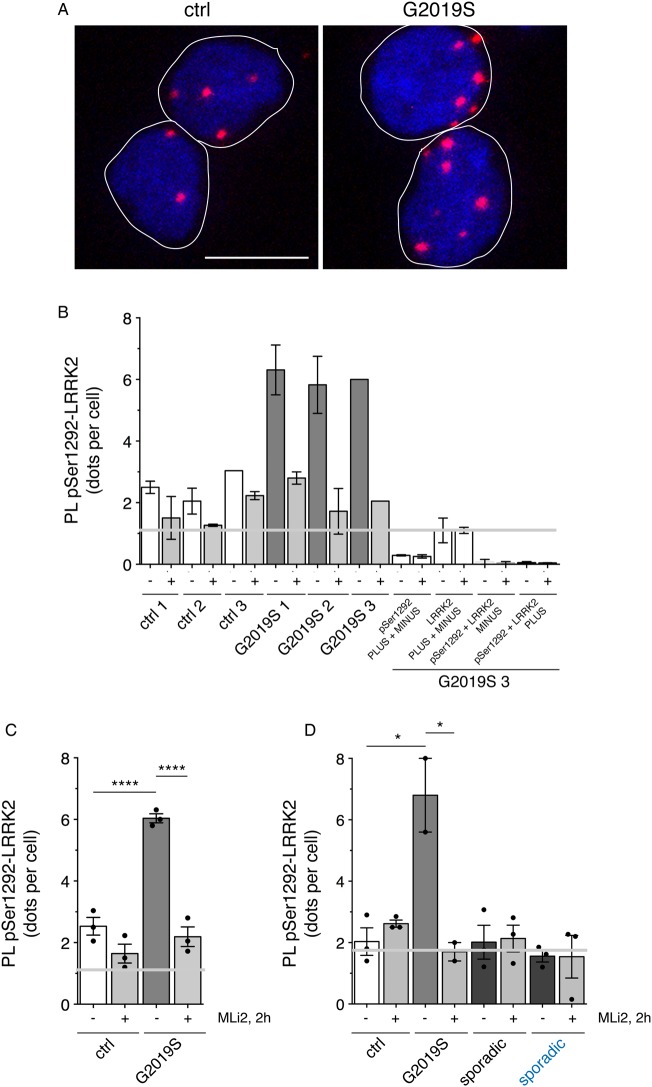
When applied to the same set of LCLs from healthy controls and G2019S LRRK2-PD patients as in Figure 1, a small phospho-Ser1292 proximity ligation signal was observed in the control samples (Figure 6A,B). G2019S LRRK2 LCLs displayed a significantly increased phospho-Ser1292 proximity ligation signal as compared with the control LCLs (Figure 6A,B), all whilst displaying very distinct levels of total LRRK2 as assessed by Western blotting techniques (Figure 1C). The proximity ligation signal was decreased upon MLi2 treatment in all three G2019S LRRK2 LCLs, indicating that it was specifically reflecting intrinsic LRRK2 kinase activity (Figure 6B,C). However, and similar to what was observed in HEK293 cells, the assay was unable to detect endogenous LRRK2 kinase activity in the control LCLs, as the signal obtained was similar to that obtained when omitting one of the two primary antibodies from the PLA reaction (Figure 6C).
Figure 6. Detection of active LRRK2 by PLAs in LCLs from control and G2019S PD patients.
(A) Example of proximity ligation signal as a readout for LRRK2 kinase activity in control and G2019S LRRK2-PD LCL cells stained for DAPI. Scale bar, 10 µm. (B) Quantification of the proximity ligation signal (PL-pSer1292-LRRK2) from three control and three G2019S LRRK2-PD LCLs in either the presence or absence of MLi2 (10 nM). Four different control proximity ligation reactions were analyzed on one G2019S LRRK2 LCL line (G2019S 3), which included omission of either one of the two primary antibodies (total LRRK2 antibody (LRRK2) or pSer1292 antibody (pSer1292)), or of either one of the two secondary antibodies (PLUS or MINUS), respectively. Note that there was significant nonspecific proximity ligation signal when omitting the pSer1292 antibody, and this signal was used as background against which the assay signals were evaluated (grey line). Around 300 cells were quantified per condition and experiment. Bars represent mean ± s.e.m. (two independent experiments). (C) Proximity ligation signal based on the means from each line as depicted in (B), from control and G2019S LRRK2 LCLs in either the presence or absence of MLi2 (10 nM) as indicated. Grey line depicts background signal when omitting the pSer1292 antibody. Bars represent mean ± s.e.m. (n = 3 independent lines); **** P < 0.001. (D) Proximity ligation signal was determined in LCLs from three distinct age-matched control, two distinct G2019S LRRK2, three sporadic PD without a cohesion phenotype, and three sporadic PD with a cohesion phenotype (blue), respectively. The nonspecific proximity ligation signal obtained when omitting the pSer1292 antibody is indicated as grey line. Bars represent mean ± s.e.m.; * P < 0.05.
Lastly, we applied the PLA to a distinct set of age-matched LCLs including three distinct controls, two distinct G2019S LRRK2 LCLs, three sporadic LCLs without a cohesion phenotype, and the three sporadic LCLs with a centrosomal cohesion phenotype, respectively (Figure 6D). Whilst the G2019S LRRK2 LCLs displayed an increased proximity ligation signal as compared with control LCLs which was reverted upon MLi2 treatment, no specific signal could be detected in either control or sporadic PD LCLs (Figure 6D). Thus, intrinsic LRRK2 kinase activity as assessed by the PLA, rather than total LRRK2 levels, seems to correlate with the observed cohesion phenotype in the G2019S LRRK2 LCLs. In contrast, the assay, at least in our hands, is unable to detect endogenous LRRK2 kinase activity in control or sporadic PD LCLs, including the sporadic PD LCLs which display a centrosomal cohesion phenotype (Figure 6D).
Finally, and given previous studies reporting the pericentrosomal/centrosomal accumulation of phospho-Rab8 and phospho-Rab10 [33,34], we wondered whether the centrosomal cohesion phenotype may correlate with altered subcellular localization of these phosphorylated Rab species. The currently available phospho-Rab8a antibodies were not of sufficient affinity to detect the pericentrosomal/centrosomal accumulation of endogenous phospho-Rab8a in LCLs (Supplementary Figure S4A). A recently developed phospho-Rab10 antibody suitable for immunocytochemistry techniques detected a pericentrosomal/centrosomal signal in some, but not all cells. When quantifying the integrated fluorescence intensity of phospho-Rab10 at/around the centrosome as defined by co-staining with a centrosomal marker, the three G2019S LRRK2 LCLs displayed a slight, but not significant increase in phospho-Rab10 accumulation as compared with control samples, which was decreased, but not abolished upon MLi2 treatment, suggesting that the antibody was not entirely phospho-state-specific, at least under the conditions employed here (Supplementary Figure S4B,C).
Discussion
The objective of this study was to evaluate whether detecting centrosomal cohesion deficits may serve as target engagement biomarker and/or patient enrichment biomarker for LRRK2 inhibitor clinical trials. All G2019S LRRK2 LCLs analyzed displayed a centrosomal cohesion deficit without alterations in cell cycle or cell viability. Such cohesion phenotype was also observed in a subset of LCLs from sporadic PD patients, and was reverted in all cases upon kinase inhibitor treatment. Thus, whilst requiring independent validation in an increased sample size of sporadic PD patients, this cellular readout may comprise a valid patient enrichment biomarker. In future experiments, it will be important to determine whether the centrosomal cohesion readout can be observed in freshly isolated PBMCs, monocytes and/or neutrophils, or whether it is dependent on lymphocyte immortalization to generate cell-cycle active LCLs. If observed in freshly isolated cells, the assay may additionally serve as a pharmacodynamic readout of potential use in LRRK2 kinase inhibitor clinical trials.
The underlying reason(s) why a subset of sporadic PD LCLs displayed a cohesion phenotype similar to G2019S LRRK2 LCLs currently remain unknown, but were not associated with increased levels of LRRK2 or Rab10 Thr73 phosphorylation as compared with healthy controls as assessed by Western blotting techniques. Whilst sequenced and found negative for the G2019S mutation in LRRK2, full sequence analysis is warranted to highlight potential other genetic alterations related to LRRK2, or indeed variations in or around other genes which are known to regulate LRRK2 activity, such as VPS35 or RAB7L1 [40–43]. In either case, these data suggest that a subset of sporadic PD patients may also benefit from LRRK2-related treatment strategies.
Previous studies in human neutrophils have reported variability between patient samples, but no consistent differences in the levels of LRRK2, LRRK2 Ser935, Rab10 or Rab10 Thr73, amongst control, G2019S LRRK2 or sporadic PD samples, respectively [31]. Similarly, we did not detect significant differences in the levels of LRRK2 and Rab10 or phospho-versions thereof between control, LRRK2 G2019S and sporadic PD LCLs, with vast differences in the levels of total LRRK2 protein amongst the distinct patient-derived LCLs. Both LRRK2 Ser935 phosphorylation and Rab10 Thr73 phosphorylation were significantly reduced upon LRRK2 inhibitor treatment in control and sporadic PD samples, with a non-significant tendency observed in LRRK2 G2019S samples. The latter is likely due to low levels of total LRRK2 observed in some LRRK2 G2019S samples, which made quantification less reliable. In either case, these data suggest that both LRRK2 Ser935 phosphorylation and Rab10 Thr73 phosphorylation are valid peripheral LRRK2 inhibition biomarkers also in LCLs, as previously described for human neutrophils and PBMCs [31,32].
Whilst there was a correlation between total LRRK2 levels and Rab10 phosphorylation levels especially in sporadic PD samples, total LRRK2 levels were not different between control and sporadic PD samples. A previous study reported an increase in LRRK2 protein levels in B cells, T cells and CD16+ monocytes from sporadic PD patients as compared with controls as assessed by flow cytometry [44]. Similarly, a recent study reported an increase in LRRK2 protein levels in neutrophils from sporadic PD patients as compared with control as assessed by Western blotting techniques [32]. Both studies employed larger patient sampling sizes, such that significant increases in LRRK2 protein levels may have been missed in the present study. In addition, the LRRK2 protein levels in immortalized lymphocytes may not reflect the LRRK2 levels present in original, freshly isolated cells. Thus, the lack of increased LRRK2 protein levels in sporadic PD LCLs as reported here is not inconsistent with the possibility that a sub-population of sporadic PD patients may benefit from being targeted with LRRK2 therapeutics.
Since differences in the intrinsic LRRK2 kinase activity, rather than total protein levels, may correlate with LRRK2-related PD, we analyzed LRRK2 kinase activity in a LCL sample set using a proximity ligation-based assay [37]. Interestingly, all G2019S LRRK2 LCLs analyzed displayed an increase in LRRK2 kinase activity as compared with controls, whilst displaying very distinct total LRRK2 levels as assessed by Western blotting techniques. However, and at least in our hands and in LCLs, the assay was not able to detect endogenous LRRK2 kinase activity in either control or sporadic PD LCLs, including the ones which displayed a centrosomal cohesion phenotype. Therefore, future studies employing improved high-affinity Ser1292 antibodies, or distinct total LRRK2 antibodies for PLAs are warranted to determine whether increased intrinsic LRRK2 kinase activity may serve as patient enrichment biomarker for sporadic PD samples, and whether there exists a correlation between LRRK2 kinase activity and the centrosomal phenotype observed in a subset of sporadic PD LCLs.
Our sporadic PD patient group displayed large differences in total LRRK2 levels and in Rab10 Thr73 phosphorylation levels which did not correlate with disease severity, disease duration or age at onset, in contrast with the reported correlation between Rab10 phosphorylation and disease severity in PBMCs from a distinct sporadic PD sampling [32]. Such lack of correlation with PD clinical variables may be due to the smaller sample size employed here, and/or the use of immortalized cells cultured ex vivo as compared with freshly isolated cells. In either case, independent studies in freshly isolated PBMCs from larger patient cohorts are required to confirm or refute a correlation between phospho-Rab10 levels and clinical PD variables.
The LRRK2 G2019S LCLs, as well as a subset of LCLs from sporadic PD patients displayed a centrosomal cohesion deficit as compared with control samples, which did not correlate with Rab10 Thr73 phosphorylation levels as assessed by Western blotting techniques. Since both phospho-Rab8 and phospho-Rab10 associate with the pericentrosomal/centrosomal RILPL1 protein [27], the centrosomal cohesion phenotype may correlate with the summed increase in both of these phospho-Rab proteins. Thus, and once phospho-state-specific antibodies uniquely selective for only phospho-Rab8a have been developed, it will be interesting to analyze for alterations in the phosphorylation states of Rab8a Thr72 by Western blotting techniques. In addition, and given the lack of a significant increase in pericentrosomal/centrosomal phospho-Rab10 fluorescence intensity in the G2019S LCLs as compared with controls, signal amplification via development of PLAs around RILPL1 and phospho-Rab8/phospho-Rab10 are warranted to determine whether the centrosomal cohesion phenotype may correlate with the pericentrosomal/centrosomal accumulation of these phosphorylated Rab species.
Control LCLs displayed phospho-Rab10 levels similar to G2019S LRRK2 LCLs, but did not show a centrosomal cohesion deficit. Application of LRRK2 kinase inhibitor to control cells caused a decrease in phospho-Rab10 levels, but no further decrease in centrosomal cohesion. Thus, the G2019S LRRK2-mediated phospho-Rab10 protein able to cause the centrosomal cohesion deficits may reflect a distinct phosphorylated protein species, driven by differences in the subcellular localization of the phosphorylation event due to differences in the subcellular localization of wildtype versus G2019S LRRK2 [40–42]. This may result in the generation of a phosphorylated Rab protein species in a distinct location and/or nucleotide-bound state [41], which may be the only one selectively able to cause the centrosomal cohesion deficits. Whilst impossible to detect by Western blotting techniques, such alterations may be sensitively detected by cell biological readouts which are due to the specific phospho-Rab species generated in the context of pathogenic LRRK2. Finally, the LRRK2 kinase activity-mediated centrosomal cohesion phenotype may also be due to the phosphorylation of other Rab protein species, and/or related to other aspects of LRRK2 biology. In either case, and whilst further studies into the mechanism(s) underlying the centrosomal deficits are required, our present data indicate that centrosomal cohesion deficits are a robust cellular readout for G2019S LRRK2 kinase activity, and may be able to stratify sporadic PD patients who may also benefit from LRRK2-related therapies.
Acknowledgements
We are grateful to L. Montosa for technical support for confocal microscopy and image analysis, S. Guerrero for flow cytometry analysis, and E. León for help with statistical analysis. We thank L. Sanders (Duke University, U.S.A.) for sharing detailed protocols related to apoptosis determination, and T. Greenamyre (University of Pittsburgh, U.S.A.) for sharing the various HEK293 cell lines as well as detailed protocols for LRRK2 proximity ligation assays.
Abbreviations
- EBV
Epstein–Barr virus
- KI
knockin
- LAS
Leica Applied Systems
- LCLs
lymphoblastoid cell lines
- LED
l-dopa-equivalent dose
- LRRK2
Leucine-rich repeat kinase 2
- MDS-UPDRS
Movement Disorders Society Unified Parkinson's Disease Rating Scale
- PBMCs
peripheral blood mononuclear cells
- PBS
phosphate-buffered saline
- PD
Parkinson's disease
- PFA
paraformaldehyde
- PLA
proximity ligation assay
Author Contribution
B.F. designed and executed the majority of experiments and analyzed data, as well as playing a major role in the preparation of most Figures for the paper. A.J.L.Ordóñez and E.F. participated in some of the experiments, including help with culturing distinct LCLs and FACS analysis. T.C. and C.L. performed lymphocyte EBV immortalization of the samples from subjects who participated in the study, and contributed to several experiments including Western blotting and immunocytochemistry, in which L.V. participated as well. E.M., C.S. and A.K. participated in the neurological evaluation of the subjects with the help of L.D., and S.B. organized samples and clinical information collection of the recruited patients under the coordination of A.D. and E.M. J.-M.T. participated in scientific discussion of the work and revised the manuscript. M.-C.C.-H. conceived part of the experimental design, selected individuals to be studied, helped in experimental interpretations and revised the manuscript. S.H. conceived the project, helped with experimental design, analysis and interpretation of the data, and wrote the paper.
Funding
This work was supported by funding from the Michael J. Fox Foundation for Parkinson's Research (U.S.A.), FEDER, the Spanish Ministry of Science, Innovation and Universities (SAF2017-89402-R), and by the French Ministry of Health's PHRC program (CONVERGENCE 2008-A00219-42), the University of Lille, Inserm and the Lille University Hospital.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
Supplementary Material
References
- 1.Kalia L.V. and Lang A.E. (2015) Parkinson's disease. Lancet 386, 896–912 10.1016/S0140-6736(14)61393-3 [DOI] [PubMed] [Google Scholar]
- 2.Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M. et al. (2014) Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet. 46, 989–993 10.1038/ng.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brás J., Guerreiro R. and Hardy J. (2015) Snapshot: genetics of Parkinson's disease. Cell 160, 570–570.e1 10.1016/j.cell.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 4.Hernandez D.G., Reed X. and Singleton A.B. (2016) Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J. Neurochem. 139, 59–74 10.1111/jnc.13593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Healy D.G., Falchi M., O'Sullivan S.S., Bonifati V., Durr A., Bressman S. et al. (2008) Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: a case-control study. Lancet Neurol. 7, 583–590 10.1016/S1474-4422(08)70117-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilks W.P., Abou-Sleiman P.M., Gandhi S., Jain S., Singleton A., Lees A.J. et al. (2005) A common LRRK2 mutation in idiopathic Parkinson's disease. Lancet 365, 415–416 10.1016/S0140-6736(05)17830-1 [DOI] [PubMed] [Google Scholar]
- 7.Bardien S., Lesage S., Brice A. and Carr J. (2011) Genetic characteristics of leucine-rich repeat kinase 2 (LRRK2) associated Parkinson's disease. Parkinsonism Relat. Disord. 17, 501–508 10.1016/j.parkreldis.2010.11.008 [DOI] [PubMed] [Google Scholar]
- 8.Trabzuni D., Ryten M., Emmett W., Ramasamy A., Lackner K.J., Zeller T. et al. (2013) Fine-mapping, gene expression and splicing analysis of the disease associated LRRK2 locus. PLoS ONE 8, e70724 10.1371/journal.pone.0070724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed X., Bandrés-Ciga S., Blauwendraat C. and Cookson M.R. (2018) The role of monogenic genes in idiopathic Parkinson's disease. Neurobiol. Dis. 124, 230–239 10.1016/j.nbd.2018.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alessi D.R. and Sammler E. (2018) LRRK2 kinase in Parkinson's disease. Science 360, 36–37 10.1126/science.aar5683 [DOI] [PubMed] [Google Scholar]
- 11.Greggio E. and Cookson M.R. (2009) Leucine-rich repeat kinase 2 mutations and Parkinson's disease: three questions. ASN Neuro 1, e00002 10.1042/AN20090007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steger M., Diez F., Dhekne H.S., Lis P., Nirujogi R.S., Karayel O. et al. (2016) Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5, e12813 10.7554/eLife.12813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taymans J.M. and Greggio E. (2016) LRRK2 kinase inhibition as a therapeutic strategy for Parkinson's disease, where do we stand? Curr. Neuropharmacol. 14, 214–225 10.2174/1570159X13666151030102847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West A.B. (2017) Achieving neuroprotection with LRRK2 kinase inhibitors in Parkinson's disease. Exp. Neurol. 298, 236–245 10.1016/j.expneurol.2017.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cookson M.R. (2017) Mechanisms of mutant LRRK2 neurodegeneration. Adv. Neurobiol. 14, 227–239 10.1007/978-3-319-49969-7_12 [DOI] [PubMed] [Google Scholar]
- 16.Nolen B., Taylor S. and Ghosh G. (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675 10.1016/j.molcel.2004.08.024 [DOI] [PubMed] [Google Scholar]
- 17.Sheng Z., Zhang S., Bustos D., Kleinheinz T., Le Pichon C.E., Dominguez S.L. et al. (2012) Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci. Transl. Med. 4, 164ra161 10.1126/scitranslmed.3004485 [DOI] [PubMed] [Google Scholar]
- 18.Nichols R.J., Dzamko N., Morrice N.A., Campbell D.G., Deak M., Ordureau A. et al. (2010) 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochem. J. 430, 393–404 10.1042/BJ20100483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dzamko N., Deak M., Hentati F., Reith A.D., Prescott A.R., Alessi D.R. et al. (2010) Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser910/Ser935, disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem. J. 430, 405–413 10.1042/BJ20100784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito G., Fujimoto T., Kamikawaji S., Kuwahara T. and Iwatsubo T. (2014) Lack of correlation between the kinase activity of LRRK2 harboring kinase-modifying mutations and its phosphorylation at Ser910, 935 and Ser955. PLoS ONE 9, e97988 10.1371/journal.pone.0097988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanca Ramírez M., Lara Ordóñez A.J., Fdez E., Madero-Pérez J., Gonnelli A., Drouyer M. et al. (2017) GTP binding regulates cellular localization of Parkinson's disease-associated LRRK2. Hum. Mol. Genet. 26, 2747–2767 10.1093/hmg/ddx161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vancraenenbroeck R., De Raeymaecker J., Lobbestael E., Gao F., De Maeyer M., Voet A. et al. (2014) In silico, in vitro and cellular analysis with a kinome-wide inhibitor panel correlates cellular LRRK2 dephosphorylation to inhibitor activity on LRRK2. Front. Mol. Neurosci. 7, 51 10.3389/fnmol.2014.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobbestael E., Zhao J., Rudenko I.N., Beylina A., Gao F., Wetter J. et al. (2013) Identification of protein phosphatase 1 as a regulator of the LRRK2 phosphorylation cycle. Biochem. J. 456, 119–128 10.1042/BJ20121772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatcher J.M., Choi H.G., Alessi D.R. and Gray N.S. (2017) Small-molecule inhibitors of LRRK2. Adv. Neurobiol. 14, 241–264 10.1007/978-3-319-49969-7_13 [DOI] [PubMed] [Google Scholar]
- 25.Perera G., Ranola M., Rowe D.B., Halliday G.M. and Dzamko N. (2016) Inhibitor treatment of peripheral mononuclear cells from Parkinson's disease patients further validates LRRK2 dephosphorylation as a pharmacodynamic biomarker. Sci. Rep. 6, 31391 10.1038/srep31391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dzamko N., Chua G., Ranola M., Rowe D.B. and Halliday G.M. (2013) Measurement of LRRK2 and Ser910/935 phosphorylated LRRK2 in peripheral blood mononuclear cells from idiopathic Parkinson's disease patients. J. Parkinson’s Dis. 3, 145–152 10.3233/JPD-130174 [DOI] [PubMed] [Google Scholar]
- 27.Steger M., Diez F., Dhekne H.S., Lis P., Nirujogi R.S., Karayel O. et al. (2017) Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. eLife 6, e31012 10.7554/eLife.31012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thirstrup K., Daechsel J.C., Oppermann F.S., Williamson D.S., Smith G.P., Fog K. et al. (2017) Selective LRRK2 kinase inhibition reduces phosphorylation of endogenous Rab10 and Rab12 in human peripheral mononuclear blood cells. Sci. Rep. 7, 10300 10.1038/s41598-017-10501-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong G.R., Jang E.H., Bae J.R., Jun S., Kang H.C., Park C.H. et al. (2018) Dysregulated phosphorylation of Rab GTPases by LRRK2 induces neurodegeneration. Mol. Neurodegener. 13, 8 10.1186/s13024-018-0240-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lis P., Burel S., Steger M., Mann M., Brown F., Diez F. et al. (2018) Development of phospho-specific Rab protein antibodies to monitor in vivo activity of the LRRK2 Parkinson's disease kinase. Biochem. J. 475, 1–22 10.1042/BCJ20170802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan Y., Howden A.J.M., Sarhan A.R., Lis P., Ito G., Martinez T.N. et al. (2018) Interrogating Parkinson's disease LRRK2 kinase pathway activity by assessing Rab10 phosphorylation in human neutrophils. Biochem. J. 475, 23–44 10.1042/BCJ20170803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atashrazm F., Hammond D., Perera G., Bolliger M.F., Matar E., Halliday G.M. et al. (2018) LRRK2-mediated rab10 phosphorylation in immune cells from Parkinson's disease patients. Mov. Disord. 34, 406–415 10.1002/mds.27601 [DOI] [PubMed] [Google Scholar]
- 33.Dhekne H.S., Yanatori I., Gomez R.C., Tonelli F., Diez F., Schuele B. et al. (2018) A pathway for Parkinson's disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain. eLife 7, e40202 10.7554/eLife.40202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madero-Pérez J., Fdez E., Fernández B., Lara Ordóñez A.J., Blanca Ramírez M., Gómez-Suaga P. et al. (2018) Parkinson disease-associated mutations in LRRK2 cause centrosomal defects via Rab8a phosphorylation. Mol. Neurodener. 13, 3 10.1186/s13024-018-0235-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassin-Baer S., Laitman Y., Azizi E., Mochadski I., Galore-Haskel G., Barak F. et al. (2009) The leucine rich repeat kinase 2 (LRRK2) G2019S substitution mutation. Association with Parkinson disease, malignant melanoma and prevalence in ethnic groups in Israel. J. Neurol. 256, 483–487 10.1007/s00415-009-0117-x [DOI] [PubMed] [Google Scholar]
- 36.Louie L.G. and King M.C. (1991) A novel approach to establishing permanent lymphoblastoid cell lines: Epstein-Barr virus transformation of cryopreserved lymphocytes. Am. J. Hum. Genet. 48, 637–638 PMID: [PMC free article] [PubMed] [Google Scholar]
- 37.Di Maio R., Hoffman E.K., Rocha E.M., Keeney M.T., Sanders L.H., De Miranda B.R. et al. (2018) LRRK2 activation in idiopathic Parkinson's disease. Sci. Transl. Med. 10, eaar5429 10.1126/scitranslmed.aar5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nigg E.A. and Stearns T. (2011) The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell. Biol. 13, 1154–1160 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y., Perera G., Takahashi-Fujigasaki J., Mash D.C., Vonsattel J.P.G., Uchino A. et al. (2018) Reduced LRRK2 in association with retromer dysfunction in post-mortem brain tissue from LRRK2 mutation carriers. Brain 141, 486–495 10.1093/brain/awx344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purlyte E., Dhekne H.S., Sarhan A.R., Gomez R., Lis P., Wightman M. et al. (2018) Rab29 activation of the Parkinson's disease-associated LRRK2 kinase. EMBO J. 37, 1–18 10.15252/embj.201798099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z., Bryant N., Kumaran R., Beilina A., Abeliovich A., Cookson M.R. et al. (2018) LRRK2 phosphorylates membrane-bound Rabs and is activated by GTP-bound Rab7L1 to promote recruitment to the trans-Golgi network. Hum. Mol. Genet. 27, 385–395 10.1093/hmg/ddx410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madero-Pérez J., Fernández B., Lara Ordóñez A.J., Fdez E., Lobbestael E., Baekelandt V. et al. (2018) Rab7L1-mediated relocalization of LRRK2 to the Golgi complex causes centrosomal deficits via Rab8a. Front. Mol. Neurosci. 11, 417 10.3389/fnmol.2018.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mir R., Tonelli F., Lis P., Macartney T., Polinski N.K., Martinez T.N. et al. (2018) The Parkinson's disease VPS35[D620N] mutation enhances LRRK2-mediated Rab protein phosphorylation in mouse and human. Biochem. J. 475, 1861–1883 10.1042/BCJ20180248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cook D.A., Kannarkat G.T., Cintron A.F., Butkovich L.M., Fraser K.B., Chang J. et al. (2017) LRRK2 levels in immune cells are increased in Parkinson's disease. NPJ Parkinsons Dis. 3, 11 10.1038/s41531-017-0010-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.