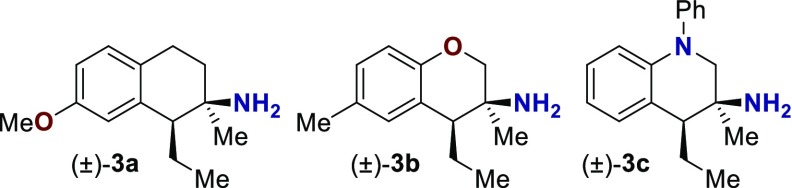
Table 1. Initial GPCR Screena.
| pKia |
|||
|---|---|---|---|
| receptor | compound 3a | compound 3b | compound 3c |
| 5-HT1A | 6.2 ± 0.1b | 5.7 ± 0.1b | 7.13 ± 0.06b |
| 5-HT1B | <5 | <5 | <5 |
| 5-HT1D | <5 | <5 | <5 |
| 5-HT1E | <5 | <5 | <5 |
| 5-HT2A | <5 | <5 | <5 |
| 5-HT2B | 6.84 ± 0.06b | 6.75 ± 0.06b | 6.1 ± 0.1b |
| 5-HT2C | 5.6 ± 1.0b | 5.6 ± 3.5b | 5.4 ± 1.3b |
| 5-HT3 | <5 | <5 | <5 |
| 5-HT5A | 5.7 ± 1.5b | <5 | <5 |
| 5-HT6 | <5 | <5 | <5 |
| 5-HT7 | <5 | <5 | 6.0 ± 0.1b |
| Alpha1A | <5 | <5 | 5.8 ± 0.1b |
| Alpha1B | <5 | <5 | <5 |
| Alpha1D | <5 | <5 | <5 |
| Alpha2A | 5.2 ± 0.2b | <5 | 6.4 ± 0.1b |
| Alpha2B | 5.3 ± 0.1b | <5 | 6.4 ± 0.1b |
| Alpha2C | <5 | <5 | <5 |
| Beta1 | <5 | <5 | <5 |
| Beta2 | 5.3 ± 0.1b | <5 | <5 |
| Beta3 | <5 | <5 | 6.0 ± 0.1b |
| BZP | 5.8 ± 0.1b | <5 | 5.1 ± 0.1b |
| D1 | <5 | <5 | <5 |
| D2 | <5 | <5 | <5 |
| D3 | <5 | <5 | <5 |
| D4 | <5 | <5 | 5.7 ± 0.1b |
| D5 | <5 | <5 | <5 |
| DAT | 5.3 ± 0.2b | <5 | 5.5 ± 0.2b |
| GABAA | <5 | <5 | <5 |
| H1 | 5.5 ± 0.5b | <5 | <5 |
| H2 | <5 | <5 | <5 |
| H3 | <5 | <5 | 5.4 ± 0.2b |
| H4 | <5 | <5 | <5 |
| M1 | <5 | <5 | <5 |
| M2 | <5 | <5 | <5 |
| M3 | <5 | <5 | <5 |
| M4 | <5 | <5 | <5 |
| M5 | <5 | <5 | <5 |
| NET | 5.6 ± 0.1b | 6.5 ± 0.1b | 6.4 ± 0.1c |
| δ-OR | <5 | <5 | <5 |
| κ-OR | 6.5 ± 0.1b | <5 | <5 |
| μ-OR | 5.7 ± 0.1b | <5 | <5 |
| PBR | <5 | <5 | <5 |
| σ1 | 6.9 ± 0.1b | 6.0 ± 0.1b | 6.5 ± 0.1b |
| σ2 | <5 | <5 | 6.2 ± 0.1b |
For pKi values reported as <5, the compound did not demonstrate ≥50% binding in an initial assay at 10 μM concentration.
These data reflect triplicate measurements of a single experiment. The error reflects the error in the sigmoidal curve fit.
These data reflect the average of triplicate experiments run in triplicate. The error reflects the standard deviation of the three calculated pKi values. Values in italic type are noted for the reader’s convenience.