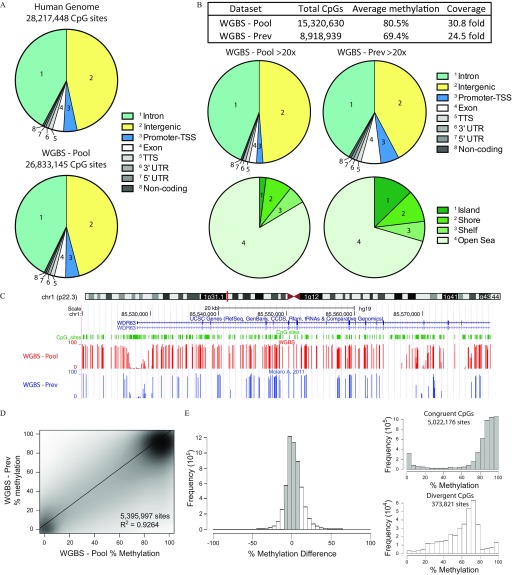
Figure 1.
Whole genome bisulfite sequencing (WGBS) of human sperm DNA. (A) Examination of the genomic distribution of CpG sites found within the human genome (top) and from WGBS (bottom) of a pooled DNA sample (WGBS-Pool). (B) Analysis of highly covered () sites from WGBS-Pool and that of WGBS-Prev examining their genomic distribution and their location within CpG islands, shores and shelves. (C) Genome browser view of DNA methylation tracks from WGBS-Pool and WGBS-Prev data sets. (D) Scatter plot of common CpGs sequenced at between WGBS data sets. (E) Difference in DNA methylation of common CpG sites (WGBS-Pool minus WGBS-Prev, left). DNA methylation of sites congruent ( methylation difference, shaded bars) and divergent ( methylation difference, open bars) sites, right top and bottom, respectively. Human genome total CpG sites were derived from the human genome sequence and downloaded through the WashU EpiGenome Browser (https://egg.wustl.edu/d/hg19/CpGsites.gz). Note: chr, chromosome; CpG, 5′-cytosine-phosphate-guanine-3′; TSS, transcriptional start site; TTS, transcriptional termination site; UTR, untranslated region; WGBS-pool, WGBS library pool; CCDS, consensus coding sequence; WGBS-Prev, publically available data on human sperm from Molaro et al. (2011).