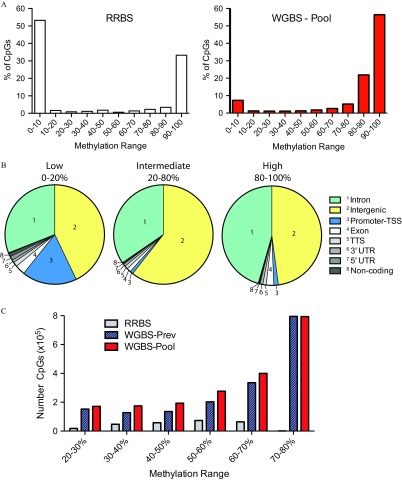
Figure 2.
Distribution of sperm DNA methylation from our WGBS-Pooled sample. (A) Percentage of CpGs with specific DNA methylation ranges sequenced from RRBS (left) the WGBS-Pool (, right). (B) Genomic distribution of CpG sites with low (0–20%), intermediate (20–80%), and high (80–100%) DNA methylation from WGBS-Pool data. (C) Number of CpG sites demonstrating intermediate levels of DNA methylation interrogated by RRBS, WGBS-Prev, and WGBS-Pool. WGBS-Prev data is from Molaro et al. 2011. RRBS data is from Aarabi et al. 2015. Note: CpG, 5′-cytosine-phosphate-guanine-3′; RRBS, reduced representation bisulfite sequencing; TSS, transcriptional start site; TTS, transcriptional termination site; UTR, untranslated region; WGBS, whole genome bisulfite sequencing; WGBS-pool, WGBS library pool; WGBS-Prev, publically available data on human sperm from Molaro et al. (2011).