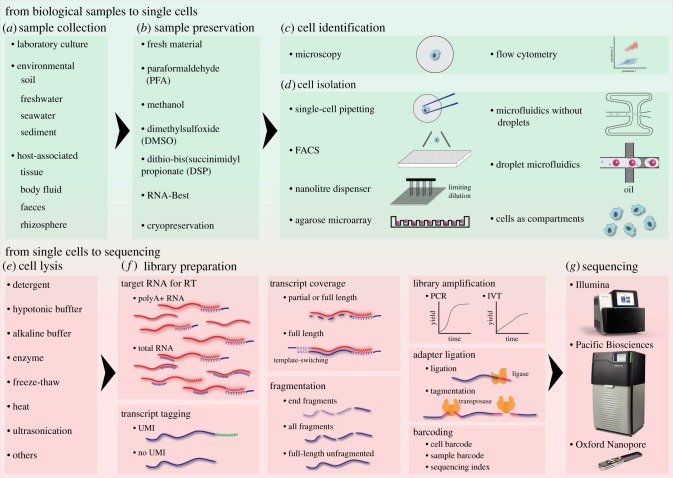
Figure 1.
Overview of the main steps and alternative strategies in single-cell RNA sequencing methods. (a,b) Biological samples taken from the laboratory or the field are either processed immediately or preserved for later use. (c,d) A variety of single-cell isolation platforms provide different throughput and possibilities to characterize individual cells. (e,f) Each single cell is usually lysed in its own compartment, and the targeted RNA is converted to cDNA, tagged with a specific cell barcode to identify the cellular origin of each RNA molecule. Various library preparation protocols offer different options for transcript quantification, coverage, and fragmentation, signal amplification, adapter ligation, and barcoding strategies, which depend on the sequencing technology (g) as well as the study design. RT, reverse transcription; UMI, unique molecular identifier. (Online version in colour.)