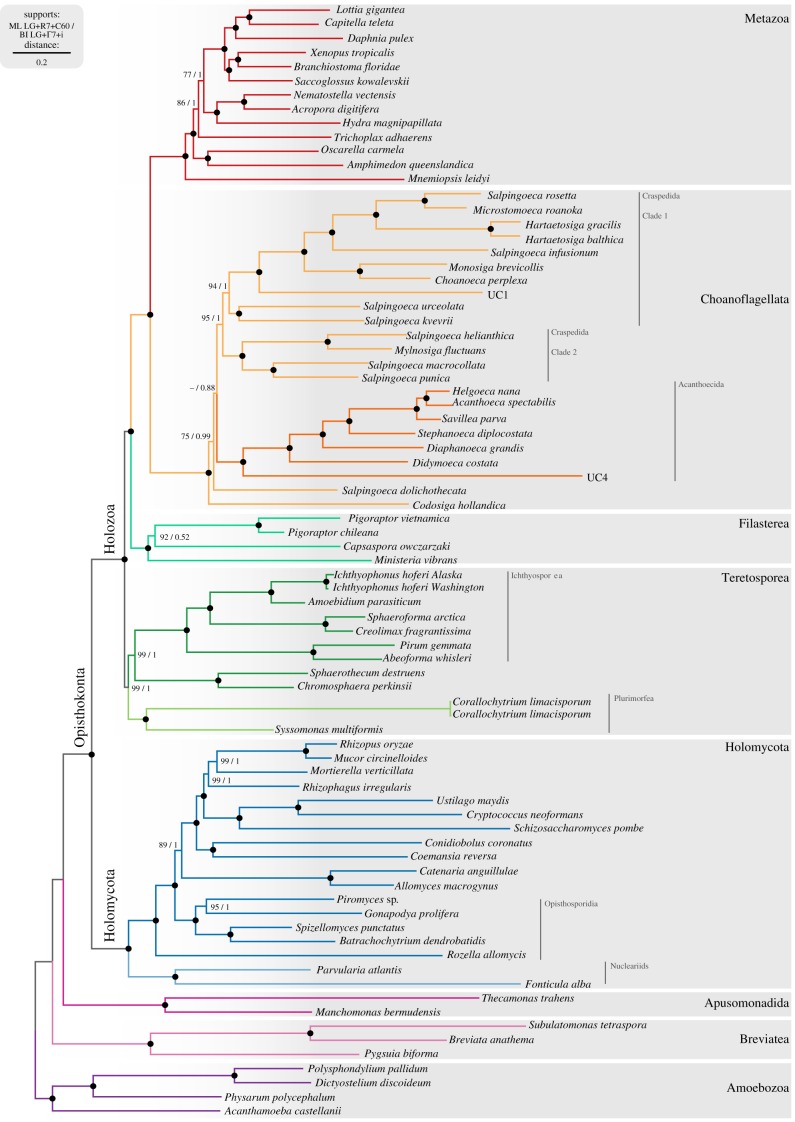
Figure 2.
Phylogenomic tree of holozoans. Phylogenomic analysis of 87 single-copy protein domains [14] accounting for 23 364 amino acid positions. Tree topology is the consensus of two Markov chain Monte Carlo chains run for 5660 and 5685 generations, after a burn-in of 13%. Statistical supports are indicated at each node: on the left, non-parametric ML ultrafast-bootstrap (UFBS) values obtained from 1000 replicates using IQ-TREE and the LG+R7+C60 model; on the right, Bayesian posterior probabilities (BPP) under the LG+Γ7+CAT model as implemented in Phylobayes. Nodes with maximum support values (BPP = 1 and UFBS = 100) are indicated with a black bullet. Raw trees are available on Figshare (https://doi.org/10.6084/m9.figshare.7819571.v1) and electronic supplementary material, figure S1 shows the topology and the supports of the ML inference.