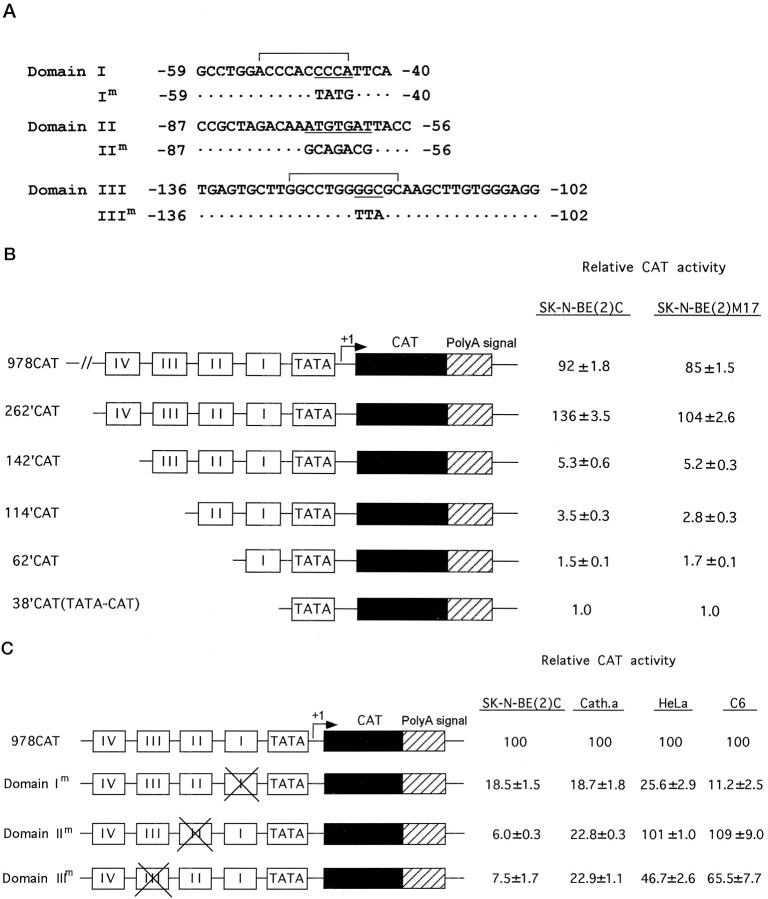
Fig. 1.
Interdependent activation of DBH transcription by proximal protein-binding sites in DBH-expressing cells.A, Nucleotide sequences and the locations of domains I, II, and III of the human DBH gene, as identified by DNase I footprint analysis. The Sp1-binding motif and AP2-binding motif residing in domain I and III, respectively, are indicated by brackets. In contrast, domain II did not show any significant sequence homology to known binding motifs except an ATTA motif at its 3′ side. Base substitutions within each domain that are analyzed by EMSA and transient transfection assays in this study are also indicated. B, Promoter activities of deletional DBH-CAT reporter constructs were determined by transient transfection assays in DBH-expressing SK-N-BE(2)C and SK-N-BE(2)M17 cell lines and expressed relative to that of the minimal 38′CAT construct. C, Effect of site-directed mutation of each cis-regulatory element on DBH promoter activity in the context of the upstream 978 bp sequences in DBH-expressing [SK-N-BE(2)C and CATH.a] and nonexpressing (HeLa and C6) cell lines. The normalized CAT activity driven by 978CAT in each cell line was set to 100 to compare the effect of each mutation on cell-specific promoter function of the DBH upstream sequence. The relative values are presented as mean ± SEM values from six to eight independent samples. Base substitutions in domain II diminished DBH promoter activity exclusively in DBH-expressing cells, virtually rendering the upstream DBH sequence a nonspecific promoter.