A CC-NBS-LRR gene underlies the Le4 locus for interspecific hybrid lethality between Gossypium barbadense and G. hirsutum, and appears to act by triggering autoimmune responses.
Keywords: ATP-binding, autoimmunity response, cotton, DNA-dependent ATPase activity, hybrid lethality, map-based cloning, NBS-LRR genes
Abstract
Hybrid lethality forms a reproductive barrier that has been found in many eukaryotes. Most cases follow the Bateson–Dobzhansky–Muller genetic incompatibility model and involve two or more loci. In this study, we demonstrate that a coiled-coil nucleotide-binding site leucine-rich repeat (CC-NBS-LRR) gene is the causal gene underlying the Le4 locus for interspecific hybrid lethality between Gossypium barbadense and G. hirsutum (cotton). Silencing this CC-NBS-LRR gene can restore F1 plants from a lethal to a normal phenotype. A total of 11 099 genes were differentially expressed between the leaves of normal and lethal F1 plants, of which genes related to autoimmune responses were highly enriched. Genes related to ATP-binding and ATPase were up-regulated before the lethal syndrome appeared; this may result in the conversion of Le4 into an active state and hence trigger immune signals in the absence of biotic/abiotic stress. We discuss our results in relation to the evolution and domestication of Sea Island cottons and the molecular mechanisms of hybrid lethality associated with autoimmune responses. Our findings provide new insights into reproductive isolation and may benefit cotton breeding.
Introduction
Hybrid incompatibility (HI) as a by-product of genetic divergence between incipient species plays an essential role in the maintenance of species identity. There are several categories of HI, including hybrid sterility, inviability, weakness, necrosis, and lethality (Bomblies et al., 2007; Rieseberg and Willis, 2007; Rieseberg and Blackman, 2010). Hybrid lethality, a type of post-zygotic hybrid incompatibility with phenotypic manifestations that include cell death, tissue necrosis, reddening, wilting, dwarfism, and reduced growth rate (Song et al., 2009), exists widely in inter- and intraspecific hybrids of eukaryotes, including animals (Provine, 1991; Sasa et al., 1998; Presgraves and Noor, 2002; Price et al., 2002) and plants such as barley, wheat, tomato, cowpea, Oryza, Mimulus guttatus, Phaseolus vulgaris, and Nicotiana (Jeuken et al., 2009; Chen et al., 2013, 2014; Ma, 2017). The basic genetic mechanisms that underpin HI are well understood. Two or more mutational differences in genes between species interact epistatically to cause hybrid inviability or sterility. This is known as the Bateson–Dobzhansky–Muller (BDM) Model (Bateson, 1909; Dobzhansky, 1937; Muller, 1942). After years of study, many genes that cause hybrid lethality and necrosis have been isolated. It has been found that most encode proteins related to disease resistance, including nucleotide-binding site leucine-rich repeat (NBS-LRR)-type resistance proteins and their interacting proteins in Arabidopsis (Bomblies et al., 2007; Alcázar et al., 2009, 2010, 2014; Chae et al., 2014), lettuce (Jeuken et al., 2009), rice (Chen et al., 2013, 2014), and Nicotiana (Ma, 2017). The majority of plant disease resistance (R) genes encode NBS-LRR proteins. Based on the presence of a Toll/interleukin-1 receptor (TIR) domain or a coiled-coil (CC) domain, NBS-LRR genes have been classified into two main groups: TIR-NBS-LRR and CC-NBS-LRR. R genes often reside in clusters and exhibit high polymorphism and copy number variations through genome duplication, gene conversion, and illegitimate recombination (Sukarta et al., 2016). However, we have less knowledge of the molecular nature of, and the forces leading to, hybrid incompatibility during evolution. Is the accumulation of these mutations caused by natural processes or by positive selection? The answer may reveal ecological advantages for plants or help to resolve intragenomic conflicts. The development of genetic tools has facilitated positional cloning, the most straightforward system for addressing the question. Most progress in this area has been made in model species such as Drosophila, Arabidopsis, and rice. However, to identify the evolutionary forces that affect HI during speciation, various species pairs within natural populations need to be explored (Zuellig and Sweigart, 2018).
The cotton genus (Gossypium) is divided into eight diploid genome groups (A–G and K), as well as one allopolyploid clade (AD genome) formed from an ancient merger and chromosome-doubling from A and D genome ancestors (Paterson et al., 2012; Wang et al., 2012). F1 necrosis and hybrid lethality have been commonly reported in Gossypium. Hybrid lethality has been reported in interspecific crosses of G. herbaceum (A1) and other species, including G. arboreum (A2) and G. anomalun (B1) (Silow, 1941). It has also been found in some F1 crosses between G. hirsutum var. marie-galante (AD)1 and G. barbadense (AD)2 (Stephens, 1946), G. hirsutum and Asiatic G. arboreum (sanguineum) (Gerstel, 1954), G. hirsutum and G. gossypioides (D6) (Phillips and Merritt, 1972), and G. klotzchianum (D3-k) and other species containing two A genomes, four E genomes, one F genome, and other D genomes (Phillips and Reid, 1975). Hybrid lethality occurs in crosses between G. davidsonii (D3-d) and G. hirsutum or G. barbadense (Lee, 1981) and the causal genes, Le2 and Le1, have been found to be located on chromosome (chr.) D12 (Stelly, 1990) and chr. A12 (Samora et al., 1994), respectively. In our previous studies, we reported a novel combination of hybrid lethality in tetraploid cotton corresponding to the BDM model (Song et al., 2009). The hybrid lethality gene Le3 exists in G. hirsutum and Le4 exists in G. barbadense acc. Coastland R4-4 (R4-4). These two genes were mapped to chr. D8 and chr. D11, respectively. In this current study, by deploying a map-based cloning strategy, a CC-NBS-LRR gene causing interspecific hybrid lethality was identified in Gossypium. Our results have implications in evolutionary biology, molecular genetics, and cotton breeding.
Materials and methods
Plant material
Gossypium barbadense acc. Coastland R4-4 (R4-4), G. barbadense cv. Hai7124, and G. hirsutum acc. Texas Marker-1 (TM-1) plants were used to produce F1 progeny. R4-4 was developed from intraspecies crossing of three G. barbadense types: Sea Island, Egyptian, and Tanguis (Jenkins, 1953). Hai7124 is the offspring of an individual selected from research into the inheritance of cotton resistance traits and is extensively grown in China. TM-1 is a standard reference cotton (G. hirsutum L.) for genetic and cytogenetic research, developed at the Texas Agricultural Experiment Station (Kohel et al., 1970). The inbred line TM-1 was used as the female parent in crosses with R4-4 and Hai7124 at the Jiangpu Breeding Station/Nanjing Agricultural University (JBS-NAU), Nanjing, China. The (TM-1×R4-4)F1 plants were backcrossed with TM-1 to produce BC1 plants on Hainan Island in China. All the resulting F1 and BC1 plants, as well as the parents, were grown for field evaluation at JBS-NAU. All plants were grown in rows spaced 80 cm apart with 30 cm between plants. Standard agronomic practices were applied from sowing until maturity. Field evaluation of (TM-1×R4-4)BC1 and (TM-1×R4-4)F1, along with their parents, was conducted in 2013 and 2015.
To further trace the origin of the gene resulting in the lethal phenotype, 46 G. hirsutum accessions (Supplementary Table S1 at JXB online) were used as female parents in crosses with R4-4 at JBS-NAU in 2009. In addition, G. barbadense acc., cv. Hai1, Giza45, VIR-4TV, BUHARA ZTW, Xinhai21, Xinhai25, Hai7124, and 3–79 were used as male parents in crosses with G. hirsutum acc. TM-1 at JBS-NAU.
Phenotypic analysis
Phenotypes were scored every 2 weeks beginning at the first true leaf stage and continuing to maturity of individual plants in the field at JBS-NAU. Phenotypes were designated as lethal or normal, depending upon the leaf appearance. Leaves of unusual appearance, characterized by reddening spots and ultimately lethal necrosis on the upper and lower surfaces, were designated as ‘lethal’, and normal green leaves as the normal phenotype. For all tests conducted in the field, F1 plants and the two parents were used as controls. The numbers of lethal and normal plants were counted in the (TM-1×R4-4)TM-1 populations in 2013 and 2015. Segregation of lethal and normal plants was examined using a chi-square test to determine whether the observed ratios fitted Mendelian genetic models at the 5% level.
Measurement of chlorophyll
In July, the fifth leaves from top of the cotton plants in the field were picked and cut into 4×5-mm strips. To extract the chlorophyll, strips from individual leaves (0.01g) were put into 50-ml tubes with 25 ml of ethanol/acetone mixture (v/v 1:1) in the dark for no more than 12 h until the strips turned white. The extract was measured at absorbance values of 470, 645, and 663 nm using a UV spectrophotometer (P-330–31 Implen, Germany). The concentrations of chlorophyll a and b were measured using the method of Arnon (1949). Each genotype was measured with three biological replications.
TEM analysis
The fifth leaves from top of plants were used for TEM analysis. Samples were taken from normal (TM-1×Hai7124)F1 and lethal (TM-1×R4-4)F1 plants grown in the field, and from lethal-phenotype recovered (TM-1×R4-4)F1 plants treated with virus-induced gene silencing (VIGS) grown in a phytotron (see below). Transverse sections of the leaf samples were fixed in 2.5% glutaraldehyde in a phosphate buffer overnight at 4 °C, then in 1% OsO4 for 2 h. The samples were dehydrated through an ethanol series and embedded in Spurr’s medium prior to ultrathin sectioning. The sections were air-dried, stained, and viewed using a Hitachi H-7650 TEM (Bio-ultrastructure Analysis Laboratory of the Analysis Center of Agrobiology and Environmental Sciences, Zhejiang University).
DNA extraction and SSR and InDel marker development
Cotton genomic DNA extraction was carried out following the method of Paterson et al. (1993). Simple sequence repeat (SSR) and insertion–deletion (InDel) markers were developed using sequences that differed between TM-1 and XinHai21 (see Supplementary Table S2).
Map-based cloning of the Le4 gene
The Le4 locus was primarily mapped to a 9.4-cM region flanked by BNL1154 and BNL3279 on chr. D11 (Song et al., 2009). Using 2013 amplified BC1 plants with additional molecular markers that were developed in this work based on the TM-1 genome sequence of G. hirsutum (Zhang et al., 2015) (Supplementary Table S2), Le4 was further mapped to a 267-kb region. The gDNA of the candidate genes was amplified from TM-1 in G. hirsutum, and R4-4, Hai7124 and Xinhai21 in G. barbadense using the primers listed in Supplementary Table S2, and the PCR products were confirmed by sequencing.
Subcellular location analysis
The complete coding sequence of the Le4 (Gh_D11G2949) gene was amplified using a K1905 primer (Supplementary Table S2) and inserted into the binary vector pBinGFP4 (Hellens et al., 2000) to produce pBinGFP4::Le4 constructs. The construct was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation (Hellens et al., 2000). The subcellular localization of Le4 was determined in Nicotiana benthamiana leaf epidermal cells by A. tumefaciens infiltration. The subcellular localization of the Le4-GFP4 fusion protein with FM 4–64 fluorescence was imaged using a Zeiss LSM 710 confocal microscope. Images were processed using the Zen 2009 software (Zeiss).
VIGS assay
We isolated a 599-bp fragment from the candidate gene Gh_D11G2949 from R4-4. This fragment had the same 587-bp sequence as Gh_D11G2950 and the same 378-bp sequence as Gh_D11G2948 (Supplementary Fig. S1), and therefore it could be used to silence all three genes in our VIGS assays. To distinguish the casual genes for Le4, a 216-bp fragment of Gh_D11G2949, a 346-bp fragment from Gh_D11G2948, and a 257-bp 3´-UTR fragment from Gh_D11G2950 were further cloned from R4-4. The products were cloned into BamH I-Xba I-digested pTRV2, generating four pTRV2-gene constructs. The vectors, pTRV1 and pTRV2-gene, were introduced into A. tumefaciens strain GV3101. More than 15 individuals of (TM-1×R4-4)F1 were infiltrated with a mixture of A. tumefaciens carrying pTRV1 and pTRV2-gene. VIGS assays were carried out independently with three replicates according to the methods described by Liu et al. (2002).
RNA extraction and sequencing
The lethal developmental process was divided into four stages (S1–S4) according to the level of lethal syndrome and age of the interspecific F1 leaves. Total RNA was isolated from developing leaves of lethal (TM-1×R4-4)F1 and normal (TM-1×Hai7124)F1 plants at the different stages using a modified CTAB-sour phenol extraction method (Jiang and Zhang, 2003). The RNA quality and purity were determined using a Qubit® 2.0 Fluorometer (Invitrogen) and a 2100 Bioanalyzer (Agilent Technologies). RNA samples from two or three biological replications were prepared for library construction and sequencing.
All cDNA libraries of 20 high-quality RNA samples were prepared following the manufacturer’s instructions using a Low-Throughput Protocol in a TruSeqTM RNA Sample Preparation Kit (Illumina). All libraries were sequenced with paired-end 100-bp reads on an Illumina HiSeq 2000 platform. Adaptors and low-quality reads were removed from further analysis. All the sequence reads have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA472432.
Transcriptome data analysis
We aligned the RNA-seq reads to the reference genome using Tophat v2.0.11, which is compatible with Bowtie2 v2.2.1. Gossypium hirsutum acc. TM-1 (Zhang et al., 2015) was used as the reference genome. Transcripts were assembled from the mapped fragments sorted by reference position using Cufflinks v2.2.0. After the assembly phase, Cufflinks quantified the expression level of each transfrag. The assembled transfrags were then merged together parsimoniously by Cuffmerge, a separate component of Cufflinks. Cuffdiff, another Cufflinks program, was used to calculate expression levels in two or more samples and to identify statistically significant differences in expression between them (Trapnell et al., 2012).
To identify differentially expressed genes (DEGs), we adopted a conservative set of criteria. We selected Cuffdiff results (Trapnell et al., 2012) with a consistent log2-fold change ≥1 or ≤–1 that were also significantly expressed with false discovery rate (FDR) corrections at the level of 0.05 and fragments per kilobase million (FPKM) values ≥1. Gene ontology and KEGG pathway enrichment analyses were performed using Plant GSEA (http://structuralbiology.cau.edu.cn/PlantGSEA/analysis.php) (Yi et al., 2013).
qRT-PCR
Real-time quantitative (qRT-)PCR analysis was performed on RNA isolated from all the lethal and normal F1 plants. cDNA was synthesized using M-MLV Reverse Transcriptase (Promega). The qRT-PCR specific primers were designed using Primer Premier 3.0 online (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) and beacon designer 8. Histone3 (AF024716) was used as an internal control. qRT-PCR was carried out on a 7500 Fast Real-Time PCR System (Applied Biosystems) in a 20-μl volume containing 100 ng of cDNA, 4 pM of each primer, and 1 0μl of AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) according to the manufacturer’s protocol. The thermal cycle conditions for PCR were as follows: 95 °C for 3 min, 40 cycles of 95 °C for 15 seconds, 60 °C for 15 seconds, and 72 °C for 30 seconds. The relative expression levels were calculated using the 2–ΔΔCT method. Each sample was measured with three replications.
ATPase activity assays
Leaves from the lethal (TM-1×R4-4)F1 and normal (TM-1×Hai7124)F1 plants at the four different development stages (S1–S4) were harvested and their ATPase activities were determined using plant ATPase activity assay kits (AngleGene). The standards were diluted to 12, 8, 4, 2, and 1 U l–1. The absorbance of each standard was measured at 636 nm (optical path=1 cm) and a standard curve was constructed. The activity of each sample corresponding to their OD636 value was measured, with three biological replications.
H2O2, POD, and SOD assays
To determine H2O2 content, and peroxidase (POD) and superoxide dismutase (SOD) activities, leaves from lethal and normal F1 plants were collected at each of the four developmental stages (S1–S4). Protein levels were quantified using the Bradford method (BioRad Quick Start Bradford assay kit). H2O2 content, and POD and SOD activities were measured with reagents provided in the Micro Hydrogen Peroxide (H2O2) Assay Kit (Solarbio) for H2O2, and the SOD Assay Kit and POD Assay Kit (both AngleGene), with three biological replications.
Results
Plant death processes during expression of interspecific F1 hybrid lethality
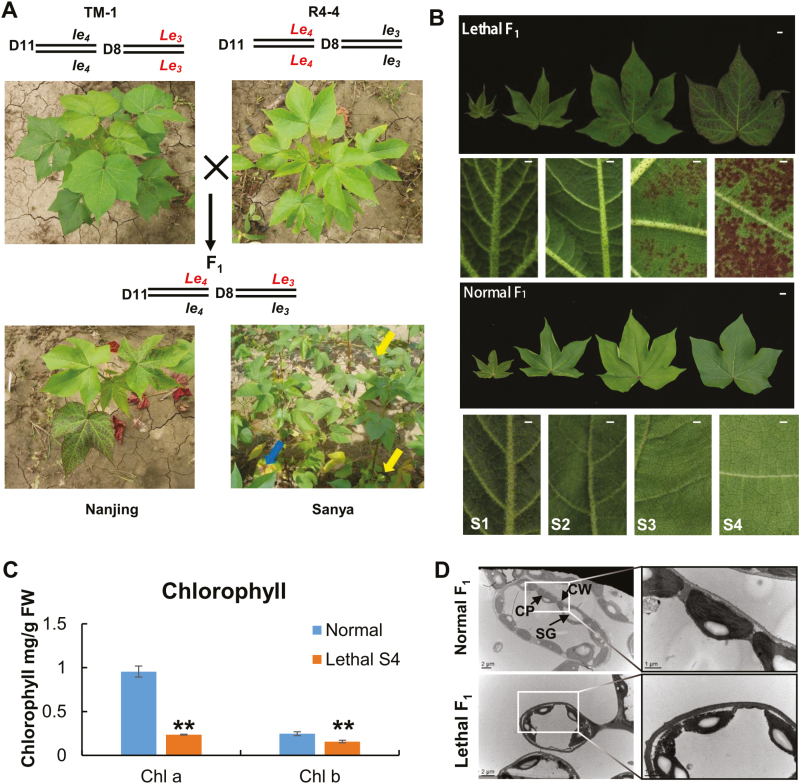
Both G. barbadense acc. R4-4 and G. hirsutum acc. TM-1 plants have normal phenotypes; however, as we previously reported (Song et al., 2009), their interspecific F1 plants exhibit a lethal phenotype with the appearance of necrotic (red) spots in leaves and infertile flowers (Fig. 1A). The lethal phenotype first appeared at the true leaf stage and remained throughout all developmental stages in the F1 plants. All newly emerging leaves initially showed the normal development phenotype, then necrotic spots arose on the abaxial side of the leaf, and gradually covered both sides, subsequently changing to red in color, before the leaves finally dropped. The lethal developmental process could be divided into four stages (S1–S4) during budding and flowering, based on the level of lethal syndrome and age of the interspecific F1 leaves (Fig. 1B). Young leaves did not display the lethal phenotype in the S1 and S2 stages, which corresponded to the development of the second and third leaves from the top of the plant, respectively. The lethal F1 leaves first displayed red necrotic spots on their adaxial and abaxial surfaces at the S3 stage, which corresponded to the fourth leaf from the top, and the intensity of the syndrome increased with the progression of leaf development until the necrotic spots had spread all over the leaf in the S4 stage, which corresponded to the fifth leaf from top. The number and size of necrotic (red) spots increased and they became increasingly severe as the leaves aged. Necrotic leaves contained less chlorophyll than normal leaves (Fig. 1C). In the lethal F1 plants, the chloroplast envelopes were blurred and the thylakoid membrane was abnormal (Fig. 1D). In addition, many cells in the lethal F1 leaves were abnormal (Supplementary Fig. S2). The symptoms could eventually lead to lethal F1 leaves being shed from the plant.
Fig. 1.
Phenotypic characterization of cotton hybrid lethality. (A) Phenotypes and genotypes of the normal parent lines TM-1 and Coastland R4-4 in the field in Nanjing, and of lethal F1 plants in the field in Nanjing and Sanya. The blue arrow indicates necrotic spots and yellow arrows indicate flower buds. D11 and D8 refer to chromosomes D11 and D08, respectively; (B) Leaves from lethal and normal F1 plants at four different developmental stages (S1–S4). The scale bars in images of complete leaves are 1 cm; in images of leaf details the bars are 0.1 cm (C) Chlorophyll content of leaves of normal and lethal F1 plants. (D) TEM images of leaves of lethal and normal F1 plants in stage S4. CP, chloroplast; CW, cell wall; SG, starch grain. Chloroplast envelopes of lethal F1 plants are blurred and the thylakoid membrane is abnormal.
The lethal F1 plants were sensitive to the environment. (TM-1×R4-4)F1 plants grown in JBS-NAU, Nanjing, Jiangsu province (32.05° N, 118.62° E), Dangtu, Anhui Province, and Kuerle, Xinjiang, China, have exhibited the lethal phenotype since we first observed them in 2004 (Song et al., 2009) (Fig. 1A). However, as previously reported (Song et al., 2009), in Sanya (18.25° N, 109.5° E), Hainan Island, China, these F1 plants show normal growth and fertility, and can be self-pollinated and/or backcrossed to produce F2 and BC1 populations for inheritance analysis and gene mapping, although some small red spots can be present in mature leaves during the full flowering stage (Fig. 1A). The cotton growing seasoning in Sanya, which runs from October to April, is under short-day control, while that in Nanjing runs from April to October and is under long-day control. Therefore, this interspecific F1 hybrid lethality may be related to photoperiod sensitivity.
A common allele in G. hirsutum interacts with Le4 to produce a lethal hybrid
To determine whether a common gene/allele in G. hirsutum interacts with the dominant gene Le4 in G. barbadense to create hybrid lethality in tetraploid cottons, crosses among a large panel of G. hirsutum and G. barbadense accessions were made and their F1 plants were screened. We first test-crossed R4-4 plants with 46 G. hirsutum accessions selected from around the world, including four major types of landraces and cultivars from the USA, as well as other domesticated subtypes from other countries (Supplementary Table S1). Without exception, all of the resulting F1 plants showed the lethal phenotype, indicating that in all the tested G. hirsutum accessions there might exist a common allele, Le3, that can complementarily interact with Le4 in R4-4 to produce a lethal hybrid phenotype, although we cannot exclude the possibility that other lethal genes exist.
Eight G. barbadense accessions representing Sea Island, American Egyptian, and Central Asia ESL types were also crossed with TM-1. With the exception of (TM-1×R4-4)F1, all F1 plants displayed a normal phenotype (Supplementary Table S3), indicating that the allele of Le4 from R4-4 is necessary for the lethality of interspecific hybrids between R4-4 and upland cotton.
Map-based isolation of the hybrid lethality gene Le4
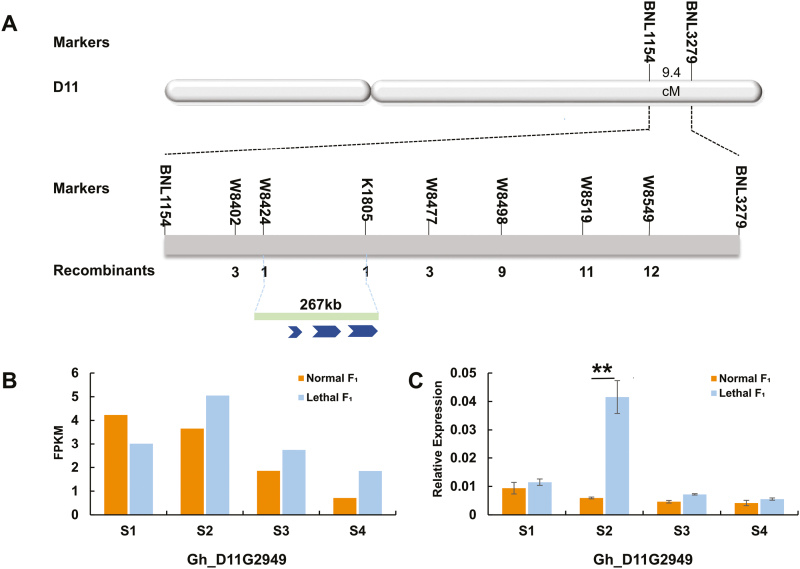
Le 4 from R4-4 was primarily anchored between two microsatellites (SSRs), BNL3279 and BNL1154, in chr. D11, with genetic distances of 5.3 cM and 4.1 cM from the Le4 allele, respectively (Fig. 2A) (Song et al., 2009). Based on the genome sequence of TM-1 (Zhang et al., 2015), Le4 was delimited to a 2.8-Mb interval on chr. D11. To fine-map this lethal-leading gene, TM-1 was crossed with R4-4 and further used as a recurrent parent to develop a large BC1 mapping population, (TM-1×R4-4)TM-1, to anchor Le4 (Supplementary Fig. S3). In 2013 and 2015, these BC1 populations comprised 1187 and 357 normal plants, and 1134 and 372 lethal plants respectively, giving a good fit to the 1:1 segregation ratio (χ21:1=1.2103 and χ21:1=0.3086). To further narrow the 2.8-Mb interval on chr. D11, a total of 150 new microsatellite primers (SSRs) and 15 InDels within in this region were developed to screen polymorphisms between R4-4 and TM-1. Le4 was thus finally delimited to a 267-kb region flanked by K1805 and W8424 (Fig. 2A) with seven SSR and InDel markers surveyed on 734 lethal individuals grown in 2013. Interestingly, all three putative ORFs (Gh_D11G2948, Gh_D11G2949, and Gh_D11G2950) were predicted to encode NBS-LRR genes according to the genomic annotation of the allotetraploid upland cotton TM-1 (Zhang et al., 2015) (Fig. 2A).
Fig. 2.
Map-based isolation of the cotton hybrid lethality gene Le4. (A) Le4 was mapped on the D11 chromosome between the markers BNL3279 and BNL1154 using an F2 population. The genetic distance between the markers was 9.4 cM. Le4 was further fine-mapped to a region between markers W8424 and K1805 using 734 dominant individuals. A 267-kb region containing three putative ORFs was obtained by mapping this region with sequences from G. hirsutum TM-1. (B) Transcriptome analysis was used to compare the expression levels of Le4 in leaves from lethal F1 and normal F1 plants at four different developmental stages (S1–S4). Data are means of two or three replicates. (C) qRT-PCR analysis was used to compare the transcript levels of Le4 in leaves from lethal F1 and normal F1 plants at four different developmental stages. Data are means (±SE) of three replicates. Significant differences between means were determined using Student’s t-test: **P<0.01.
We isolated the DNAs of these three putative genes from two interspecific mapping parents, R4-4 and TM-1. Full-length genomic sequence comparisons (Supplementary Fig. S4) revealed differences in the genomic sequences of Gh_D11G2949 and Gh_D11G2950, but not Gh_D11G2948, suggesting that Gh_D11G2949 and Gh_D11G2950 may be responsible for the production of this hybrid lethality. Single-nucleotide polymorphisms (SNPs) and InDels were identified in Gh_D11G2950 between R4-4 and TM-1; however, only two synonymous SNPs were identified in Gh_D11G2950 between the lethal-leading parent R4-4 and the wild-type, G. barbadense cv. Xinhai21 (Supplementary Fig. S5). In Gh_D11G2949, which has a full length of 3599 base pairs, 14 SNPs were found between R4-4 and TM-1, six of which were in exons and eight in a single intron. However, only three SNPs were found between R4-4 and Xinhai21. Of these, two were in introns and one was a synonymous mutation in an exon (915 G/A) that caused no difference in amino acid sequence between these two G. barbadense accessions (Supplementary Fig. S6).
The spatiotemporal expression of these three candidate genes was surveyed in the leaves of normal and lethal F1 plants. Transcriptome analysis showed that neither Gh_D11G2948 nor Gh_D11G2950 were expressed in the lethal F1 plants at any of the four development stages. However, Gh_D11G2949 was expressed, not only in all development stages in the lethal F1 plants, but also differentially in the S2 stage between the normal (TM-1×Hai7124)F1 and the lethal F1 plants (Fig. 2B). A quantitative RT-PCR (qPCR) assay of the global gene expression at the four different development stages revealed that Gh_D11G2949 had significantly higher expression levels in the lethal F1 plants than the normal F1 plants at the S2 stage (Fig. 2C). Integrated map-based cloning results implied that Gh_D11G2949 was most likely the gene responsible for hybrid lethality.
Silencing of Gh_D11G2949 leads to a normal phenotype in lethal F1 plants
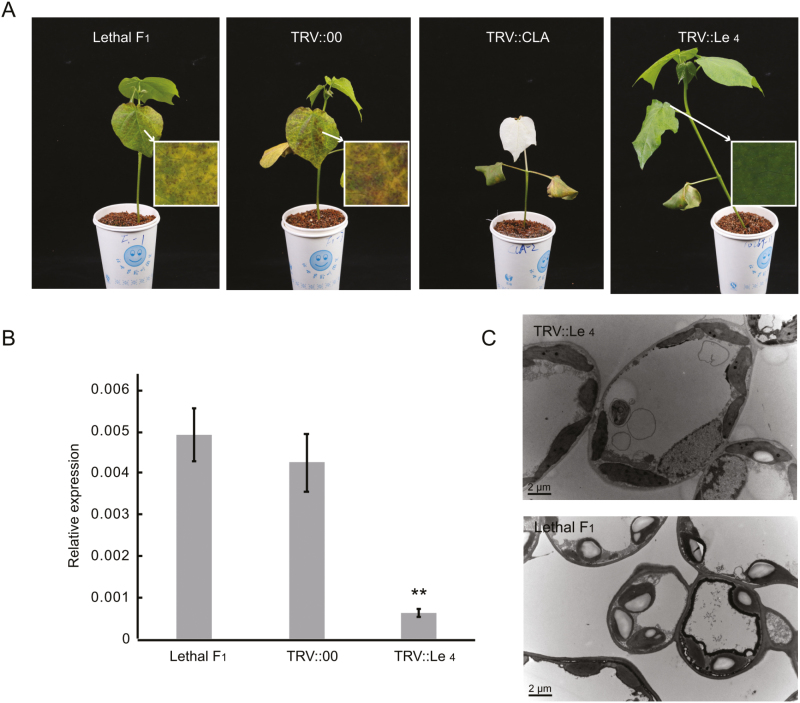
To determine whether the three NBS-LRR genes were responsible for autonomous plant death in (R4-4×TM-1)F1, a fragment (599-bp) of Gh_D11G2949 from R4-4 was isolated and inserted into pTRV2 (Qu et al., 2012) to suppress the endogenous expression of all three genes in the lethal (TM-1×R4-4)F1 plants via VIGS, since this fragment has the same sequence as the other two genes. Silencing all three of the NBS-LRR genes restored the emerging tissues of (TM-1×R4-4)F1 plants to the normal phenotype at 14 d post-agro-infiltration. No necrotic spots were visible on the lower or upper surface of leaves, and the plants were larger than those of untreated lethal F1 plants, suggesting that one of these NBS-LRR genes conferred the lethal phenotype.
To distinguish which NBS-LRR gene was responsible for the lethal phenotype, we further constructed three vectors to individually suppress the expression of the three genes (Supplementary Table S4). Firstly, in a comparison of the DNA sequences of the three genes it was found that Gh_D11G2949 had a 216-bp sequence (1162–1377) that differed to that in Gh_D11G2948 and Gh_D11G2950 (Supplementary Fig. S1). We cloned this fragment from R4-4 and inserted it into pTRV2 for VIGS of endogenous Gh_D11G2949. The emerging leaves of Gh_D11G2949-silenced (TM-1×R4-4)F1 plants had a normal phenotype at 14 d post-agro-infiltration (Fig. 3A). The transcript levels of Gh_D11G2949 in leaves of silenced plant were significantly lower than those of the untreated lethal hybrid F1 (Fig. 3B). The mesophyll cell size, and the chloroplast number and ultrastructure of the (TM-1×R4-4)F1Gh_D11G2949-silenced plants were similar to those of the normal plants (Fig. 3C). The structural and functional abnormalities compared to normal plants were thus absent in the (TM-1×R4-4)F1Gh_D11G2949-silenced plants. We further cloned 346-bp and 257-bp 3´-UTR fragments from Gh_D11G2948 and Gh_D11G2950, respectively, into R4-4, and inserted them into pTRV2 to suppress endogenous expression in (TM-1×R4-4)F1 plants. Unlike the Gh_D11G2949-silenced plants, the phenotypes of these two NBS-LRR gene-silenced lethal F1 plants were not restored to normal, indicating that Gh_D11G2948 and Gh_D11G2950 do not cause hybrid lethality. These results demonstrated that Gh_D11G2949 is the causal gene for Le4-induced autonomous plant death in (TM-1×R4-4)F1 plants.
Fig. 3.
Phenotypes of virus-induced gene silencing (VIGS) plants and suppression of endogenous transcripts in cotton. (A) Phenotypes of lethal F1 (TM-1×R4-4) plants. Detailed images of the leaves are at 25× magnification. (B) qRT-PCR analysis was used to examine the transcript levels of Le4 in leaves from plants infected with pTRV2 or pTRV2- Le4. Data are means (±SE) of three replicates. Significant differences between means were determined using Student’s t-test: **P<0.01. (C) TEM images of leaves of lethal and VIGS-silenced F1 plants.
Gh_D11G2949 is a member of the NBS-LRR gene family and contains an N-terminal coiled-coil (CC) domain, and P-loop and transmembrane regions, and therefore belongs to the CC-NBS-LRR subgroup (Supplementary Fig. S7). Transient expression of pBin-35S::Le4-GFP4 (pBin-35S::Gh_D11G2949-GFP) in N. benthamiana was co-localized in the cell membrane of transformed plants with the cell-membrane marker Fm4-64, revealing that this gene encodes a membrane protein (Supplementary Fig. S8).
Le 4 triggers autonomous plant death in interspecific hybrids
The R genes and their related causal genes can trigger autoimmunity and induce lethality or weakness (Bomblies et al., 2007; Alcázar et al., 2009, 2010, 2014; Jeuken et al., 2009; Chen et al., 2013, 2014; Chae et al., 2014; Ma, 2017). To gain insights into the regulatory circuits that control hybrid lethality, we conducted comparative transcriptome analysis between the lethal and the normal F1 leaves at four developmental stages (S1–S4) (Fig. 2B, Supplementary Table S5). Among the 70 478 genes in tetraploid cotton (Zhang et al., 2015), 11 099 exhibited 2-fold or more differential expression (FDR corrections at the level of 0.05) in the leaves in at least one stage between the lethal (TM-1×R4-4)F1 and the normal (TM-1×Hai7124)F1 plants(Supplementary Table S6). The largest number of differences in expression between the normal and lethal F1 plants occurred during the S2 stage, which corresponded to their differential expression of Le4 (Supplementary Table S7). Because Le4 expression peaked at S2 in the lethal F1 plants, and was considerably higher than in the normal F1 plants, it may act as a positive effector to trigger autoimmunity in the lethal hybrid. These results suggested that extensive transcriptome changes in the early developmental stages of the lethal F1 plants determined the fate of lethality. We therefore focused on genes that were differentially expressed in the early stages (S1 and S2) of development.
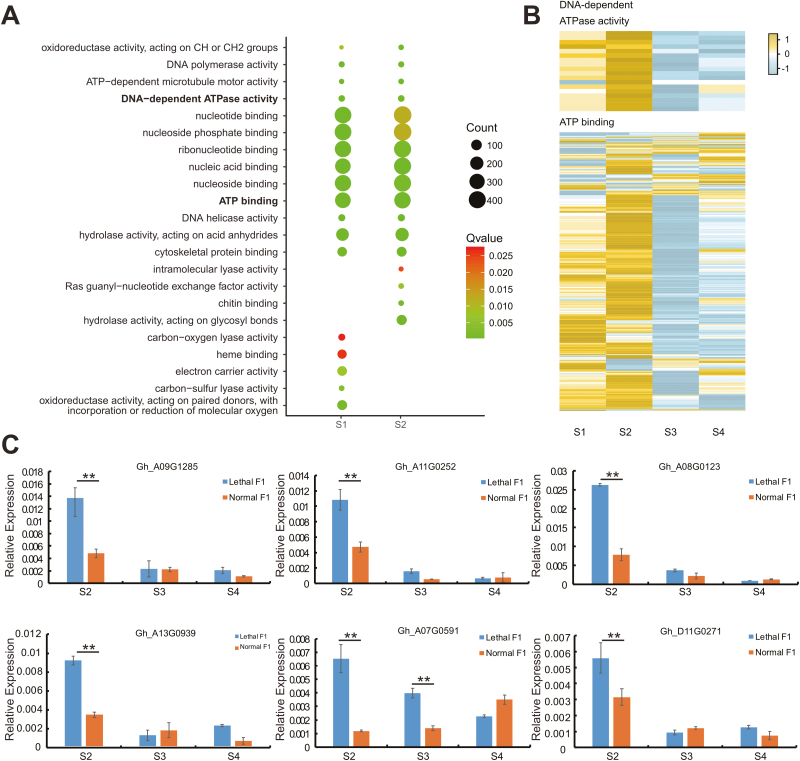
Gene ontology (GO) analysis showed that genes involved in ATP binding and DNA-dependent ATPase activity were enriched in both S1 and S2 (Fig. 4A, Supplementary Table S8). Detailed examination indicated that these two types of genes were mainly up-regulated in the early stages, and that most were not expressed or were down-regulated in S3 and S4 (Fig. 4B). To validate the transcriptome expression results, we conducted qPCR analysis of the expression levels of six representative DEGs related to ATPase activity and ATP-binding in three different development stages. The qPCR results confirmed that these genes were highly expressed at early stages in the lethal F1 leaves (Fig. 4C). In addition, in accordance with the results of the transcriptome analysis, the leaves of lethal F1 plants had higher ATPase activity in early stages than the normal F1 (Supplementary Fig. S9). ATPase activity and ATP binding are necessary for the activation of CC-NBS-LRRs (Bonardi et al., 2012; El Kasmi et al., 2017). Thus, we assumed that high expression of these components provides an appropriate environment for Le4 to change its conformation and induce autoimmunity in the early developmental stages. In S3 and S4, when the two types of genes were down-regulated or not expressed differently, the environment did not favor Le4 activation in the lethal F1 plants.
Fig. 4.
DNA dependent ATPase activity and ATP binding genes were highly expressed during developmental stages S1 and S2 in lethal F1 plants compared with normal F1 plants. (A) Enriched molecular function GO terms in up-regulated genes in the lethal F1 plants at S1 and S2. (B) Heatmap of the fold-change in expression of DNA-dependent ATPase and ATP-binding genes in lethal F1 plants. Yellow/brown indicates genes up-regulated in lethal F1 plants, blue/grey indicates genes down-regulated in lethal F1 plants, and white indicates genes with no difference in expression between lethal and normal F1 plants. (C) qPCR confirmation of the six differentially expressed ATP binding genes. Data are means (±SE) of three replicates. Significant differences between means were determined using Student’s t-test: **P<0.01.
The higher expression of these ATPase activity and ATP-binding genes related to autoimmunity induction was also confirmed in rice (Oryza sativa). Through analysis of public RNA-seq data, we found that ‘ATP binding’ (GO:0005524), ‘ATPase activity’ (GO:0016887), and ‘ATP activity, coupled’ (GO:0042623) genes were also enriched in rice at 3 h and 12 h post-inoculation (hpi) with Xanthomonas oryzae (Xo) carrying Xa7-mediated resistance-inducing protein under normal temperature conditions, compared with mock-inoculated controls (Cohen et al., 2017) (Supplementary Table S9). In this case, stronger resistance was associated with high temperature. Under high-temperature conditions, genes related to ‘ATP binding’, ‘ATPase activity’, and ‘ATPase activity, coupled’ were enriched only at 3 hpi, suggesting that stronger resistance is associated with earlier gene enrichment.
NBS-LRRs are responsible for pathogen perception (Eckardt, 2017). When CC-NBS-LRRs are activated, they trigger a downstream signaling pathway that differs to that of the TIR-NBS-LRR pathway (Toll/interleukin-1 receptor-like domain, nucleotide binding site, leucine-rich repeat). Non-race-specific disease resistance 1 (NDR1) may play an important role in the hybrid lethality that follows these signaling events (Knepper et al., 2011). We found that NDR1-like 3 was also up-regulated between S2 to S4 (Supplementary Fig. S10). Furthermore, a total of 23 mitogen-activated protein kinase (MAPK), MAP kinase kinase (MAP2K), and MAP kinase kinase kinase (MAP3K) genes were differentially expressed between the normal and the lethal F1 plants, most of which were up-regulated at stages S3 and S4 in the lethal F1 plants (Supplementary Fig. S11) after Le4 was activated. Signal transmission is mediated by a network of interacting proteins known as the MAPK signaling cascade. This suggested that immune signals are diverted by the MAPK signal pathway without biotic/abiotic stress in the lethal F1 plants. qPCR analysis of two randomly chosen MAPK/MPA2K/MAP3K genes provided results consistent with those of the transcriptome analysis (Supplementary Fig. S12). In addition, 52 WRKY (Supplementary Fig. S13) (Kim et al., 2006; Zhang et al., 2010; Niu et al., 2012; Yan et al., 2014), 13 MYB transcription factors (TFs) (Supplementary Fig. S14) (Dubos et al., 2010), and 106 pathogen-related genes (Supplementary Table S10) reportedly related to the immune response exhibited substantially higher expression in the lethal F1 plants. All these data support the suggestion that an extensive immune response was activated in lethal F1 plants in the S3 and S4 stages.
H2O2 content, and POD and SOD activities were also compared in the two F1 hybrids at each of the four stages. H2O2 is the most stable form of reactive oxygen species (ROS) (Waszczak et al., 2018), and the lethal F1 plants had higher H2O2 content and higher POD activity than the normal F1 plants in the early stages (Supplementary Figs S15, S16); however, there were no significant differences in SOD activity (Supplementary Fig. S17).
Discussion
Molecular mechanism of NBS-LRR induction of autoimmune responses
Several genes that cause hybrid lethality/necrosis/weakness have been cloned. Most of them encode common immune receptor family proteins (NBS-LRRs) or immune-related proteins in plants (Bomblies et al., 2007; Alcázar et al., 2010, 2014; Chae et al., 2014, 2016; Chen et al., 2014; Cui et al., 2015; Li et al., 2015; Świadek et al., 2017). Both DANGEROUS MIX1 (DM1) and DM2 belong to the NBS-LRR gene family and they can cause hybrid lethality in F1 plants derived from crossing Arabidopsis UK-1 and UK-3 (Chae et al., 2014). DM1/DM2d triggers hybrid lethality but requires a functional P-loop in both genes (Tran et al., 2017). RPM1 interacting protein 4 (RIN4) is a key gene in the defense response pathway that causes hybrid lethality in an interspecific lettuce hybrid (Jeuken et al., 2009). Hybrid weakness i1 (Hwi1), which encodes an LRR-RLK protein, causes hybrid lethality in rice (Chen et al., 2014). In our present study, we provide evidence that Le4, which encodes a CC-NBS-LRR protein based on the N-terminal domain, is the causative gene for the interspecific hybrid lethality between G. hirsutum and G. barbadense acc. R4-4. Like that of previously reported cases of hybrid lethality (Bomblies et al., 2007; Jeuken et al., 2009; Chen et al., 2013, 2014), CC-NBS-LRR is also an R gene and plays an important role in plant immunity (Zhang et al., 2017), but this is the first time that it has been reported to cause hybrid lethality in plants.
NBS-LRR families are expanded and enriched in many plant species (Shao et al., 2016). Le4 is located within a tandemly arrayed NBS-LRR gene cluster. Although the first plant NBS-LRR genes were cloned about 20 years ago, the functional mechanism of action of most of them is unknown (Kourelis and van der Hoorn, 2018). NBS-LRR activation results in an elevated immune response, characterized by the activation of defense-associated genes and the generation of ROS and hypersensitive responses (Spoel and Dong, 2012). In our present study, we found that autoimmunity was associated with hybrid lethality, as has been reported in a number of previous studies (Hatano et al., 2012; Chen et al., 2014; Zhang et al., 2014). Based on our experimental and transcriptome data, the mechanism for induction of autoimmunity and plant death by Le4 in the interspecific F1 hybrid between R4-4 and upland cotton is illustrated in Fig. 5. Biochemical evidence supports the idea that activation of plant NBS-LRRs is determined by the nature of the nucleotide bound to the NB-ARC domain, with the OFF and ON states being bound to ADP and ATP, respectively (Maekawa et al., 2011; Harris et al., 2013; Danot, 2015; Bentham et al., 2016; Bernoux et al., 2016). Bernoux et al. (2016) reported an equilibrium-based switch model where NBS-LRRs cycle between an inactive/ADP-bound (OFF) and active/ATP-bound (ON) state, which strongly favors the OFF state, and effector recognition shifts this equilibrium toward the ON state to activate defense signaling. We observed that genes related to ATP-binding and DNA-dependent ATPase were up-regulated in early developmental stages (Fig. 4), which could promote Le4 conversion from the OFF to the ON state, thus generating immune signals without an effector. This is the first time this equilibrium-based switch model has been used to explain the hybrid lethality phenomenon. Similar results have been found in responses of rice to X. oryzae infection (Cohen et al., 2017), providing supporting evidence for this hypothesis and indicating that it may apply not only in different species, but also in plant immune responses and hybrid incompatibility.
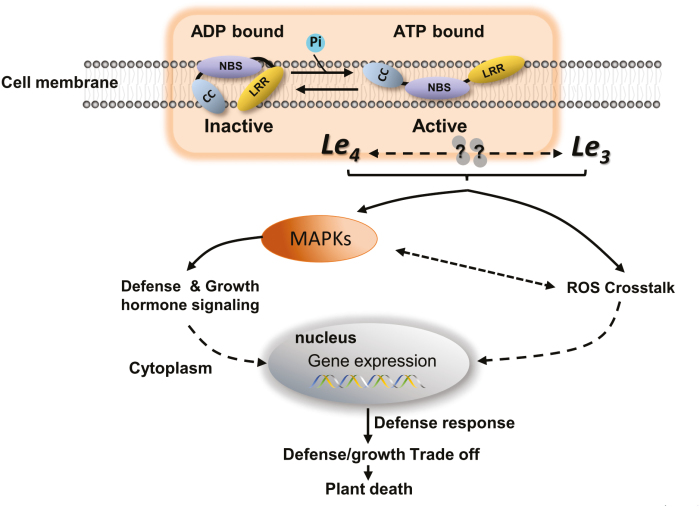
Fig. 5.
A proposed model of hybrid lethality between G. barbadense and G. hirsutum. Normally, Le4 can cycle through ON and OFF states, and favors the OFF state. When it is bound to ATP, Le4 is activated and changes from OFF to ON. Le4 interacts with Le3 directly or indirectly and triggers an immune response. Immune signals transmitted by reactive oxygen species (ROS) crosstalk and MAPK signaling pathways integrate with defense and growth hormone signals, leading to crosstalk and amplification of downstream genes. Constitutively activated defense responses result in the suppression of plant growth and ultimately induce plant death.
Due to a lack of information on Le3, we we not able to determine how it interacts with Le4 in the lethal F1. Based on our genetic analysis, both Le4 and Le3 appear to be required for hybrid lethality (Song et al., 2009). In the example of a DM1/DM2d hybrid lethality, DM1 was found to be the primary signal transducer and DM2d triggers activation of a DM1 complex via heteromerization (Tran et al., 2017), and thus the two NBS-LRR genes have unequal contributions to the activation of the immune signal . In another example involving Hwi1/Hwi2 (hybrid weakness i2), both genes appear to function in defense responses (Chen et al., 2014), and Hwi2 might act as an internal signal that is perceived by Hwi1, thus activating a downstream defense response. Canonical NBS-LRRs act as receptors that specifically recognize the presence of pathogen effectors (Zhang et al., 2017). Depending on their signaling domain, NBS-LRRs have parallel signaling pathways. TIR-NBS-LRRs require enhanced disease susceptibility 1 (EDS1), which is a lipase-like protein, whilst CC-NBS-LRRs often require NDR1, which is localized in the plasma membrane and is linked to the initiation of the MAPK pathway (Aarts et al., 1998; Knepper et al., 2011). In our study NDR1 was slightly up-regulated in S2 stage of development and significantly up-regulated in S3 and S4 (Supplementary Fig. S10). We speculate that in G. barbadense and G. hirsutum hybrids, Le3 functions as a transductor that is perceived by the highly expressed Le4, which then activates/enhances downstream defense responses. The MAPK signaling pathway, which is one of the most important singling cascades related to abiotic/biotic stimuli, transmits signals through the plasma membrane into the cell interior (Fig. 5), as also occurs in animals (Huveneers et al., 2007). ROS are important secondary messengers in core signaling pathways in plants, and maintain normal metabolic fluxes and carry out different cellular functions. Both ROS and MAPKs regulate each other’s activities, and ROS may act upstream or downstream of MAPKs (Jalmi and Sinha, 2015). Le3 and Le4 interact with each other, activating an autoimmune response in a specific genome background. This is supported by our findings in this study. Because ROS are difficult to quantify reliably in plant tissue extracts, H2O2 is commonly measured because it is the most stable of the primary ROS (superoxide, H2O2, hydroxyl radical, and singlet oxygen) compounds and it is probably the only one that is considered to be quantifiable after direct extraction (Noctor et al., 2016). In normal plants, we found that the H2O2 content did not change significantly over the four development stages, while POD activity gradually increased (Supplementary Figs S15, S16). The H2O2 content largely depends on the presence and activity of dedicated ROS scavengers (Waszczak et al., 2018), and POD functions in detoxification of H2O2 (Takahama and Oniki, 1997). Our data were consistent with the normal ageing process of leaves. Lethal F1 individuals had higher H2O2 concentrations than normal F1 in the early stages (Supplementary Fig. S15) and higher expression of MAPK-related genes (Supplementary Fig. S12). These immune signals regulate downstream signaling, defense responses, and growth hormone signaling (Eckardt, 2017) (Fig. 5). The cost of immunity as an inescapable consequence of metabolic expenditure (Huot et al., 2014) is a trade-off that ultimately induces plant death (Fig. 5).
Autoimmunity as a potential gene-flow barrier in plant species
Sea Island cottons (day-neutral forms of G. barbadense) were grown on the south-eastern seaboard of North America in the mid-eighteenth century (Stephens, 1976); however, exceedingly late maturation and low productivity in comparison with their Pima and Egyptian antecedents resulted in the cessation of their cultivation. To improve extra-long staple (ELS) cotton production, another ELS variety, Coastland cotton, was developed by crossing Sea Island, Tanguis, and Egyptian sources. However, were not produced in quantity because of their low commercial value (Jenkins, 1953). Coastland cottons such as R4-4 remained distinct during domestication and evolution. Genome resequencing data have confirmed that R4-4 is more primitive than wild or landrace species (Fang et al., 2017). This may be why we found reproductive isolation and speciation between the Coastland and G. hirsutum species, since their interspecific hybrids are lethal. However, it is very interesting that these interspecific lethal hybrids have an almost normal phenotype in Sanya, Hainan Island, China, under a short-day growing season (Song et al., 2009), similar to that of species indigenous to coastal areas of Peru, such as the landrace Tanguis. Whether or not a relationship exists between photoperiodic sensitivity and hybrid lethality in G. hirsutum needs further study.
With the spread of these two cultivated species, opportunities for crossing between them are frequent. The two presently cultivated allotetraploid species of cotton cross freely, giving vigorous and fertile F1 hybrids. One plausible explanation for the low incidence of hybrid lethality in the tetraploid cottons that are currently grown is their narrow genetic background, which has resulted from selection pressure imposed during the process of domestication and breeding, as well as long-term artificial hybridization during the movement of G. hirsutum wild relatives northward through inland Mesoamerica from the Yucatan peninsula to the Caribbean Islands, where cultivated G. barbadense originally formed.
Further molecular and functional analysis of Le4 will advance our understanding of the molecular mechanisms that underlie hybrid lethality and provide us with an opportunity to compare the regulation of this phenomenon in different species.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Alignment of the sequences of Gh_D11G2948, Gh_D11G2949, and Gh_D11G2950.
Fig. S2. TEM images of leaves of lethal and normal F1 plants in the S4 stage.
Fig. S3. Genotypes of the BC1 population.
Fig. S4. Alignment of the sequences of Gh_D11G2948 between G. barbadense R4-4 and G. hirsutum TM-1.
Fig. S5. Alignment of the sequences of Gh_D11G2950 between G. barbadense R4-4, parent line Xinhai21, and G. hirsutum TM-1.
Fig. S6. Alignment of the sequences of Gh_D11G2949 between G. barbadense R4-4, parent line Xinhai21, and G. hirsutum TM-1.
Fig. S7. Protein coding sequence and protein domain structure of Gh_D11G2949.
Fig. S8. Subcellular localization of Le4 (Gh_D11G2949) in N. benthamiana leaf epidermal cells.
Fig. S9. ATPase activity in leaves of normal and lethal F1 plants at four different developmental stages.
Fig. S10. FPKM values for the NDR1/H1N1-like 3 gene in TM-1×Hai7124 and TM-1×R4-4 at four different developmental stages.
Fig. S11. Heatmap of differentially expressed MAPK/MAP2K/MAP3K genes at four different developmental stages.
Fig. S12. qPCR of MAPK genes in leaves of normal and lethal F1 plants at four different developmental stages.
Fig. S13. Heatmap of differentially expressed WRKY transcription factors at four different developmental stages.
Fig. S14. Heatmap of differentially expressed MYB transcription factors at four different developmental stages.
Fig. S15. H2O2 concentration in leaves of lethal and normal F1 plants at four different developmental stages.
Fig. S16. Activity of POD in leaves of lethal and normal F1 plants at four different developmental stages.
Fig. S17. SOD activity in leaves of normal and lethal F1 plants at four different developmental stages.
Table S1. Phenotypes of F1 progeny of G. hirsutum accessions crossed with G. barbadense Coastland R4-4.
Table S2. Primers used in this study.
Table S3. Phenotypes of F1 progeny of G. barbadense accessions crossed with G. hirsutum TM-1.
Table S4. Summary data from four VIGS experiments.
Table S5. Statistical analysis of Tophat2 alignment of transcriptomic data from normal and lethal F1 plants.
Table S6. List of 11 099 common differentially expressed genes between the hybrid-lethal (TM-1×R4-4)F1 and the normal (TM-1×H7124)F1.
Table S7. Summary of differentially expressed genes between normal and lethal F1 plants at four different developmental stages.
Table S8. Enriched molecular function GO terms in up-regulated genes in the lethal F1 plants in early developmental stages (S1, S2).
Table S9. Enriched molecular function GO terms in up-regulated genes in rice inoculated with Xanthomonas oryzae.
Table S10. List of differentially expressed pathogen resistance genes between normal and lethal F1 plants.
Acknowledgements
This work was financially supported in part by grants from the National Key R & D Program for Crop Breeding (2016YFD0100505), the earmarked fund for the China Agriculture Research System and the Distinguished Discipline Support Program of Zhejiang University, and the Science Technology and Achievement Transformation Project of the Xinjiang Production and Construction Corps (2016AC02). The authors declare that they have no competing interests.
Author contributions
TZ conceptualized and coordinated the project; JD, XZ, and BZ planted and scored the phenotypes in Sanya and Nanjing; JD carried out mapping and cloning of Le4; JD and LF carried out the validation of Le4 through VIGS; JD and LF conducted the bioinformatic analysis of RNA-seq data; JD, TZ, and LF wrote the manuscript.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. 1998. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proceedings of the National Academy of Sciences, USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcázar R, García AV, Kronholm I, de Meaux J, Koornneef M, Parker JE, Reymond M. 2010. Natural variation at Strubbelig Receptor Kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nature Genetics 42, 1135–1139. [DOI] [PubMed] [Google Scholar]
- Alcázar R, García AV, Parker JE, Reymond M. 2009. Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proceedings of the National Academy of Sciences, USA 106, 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcázar R, von Reth M, Bautor J, Chae E, Weigel D, Koornneef M, Parker JE. 2014. Analysis of a plant complex resistance gene locus underlying immune-related hybrid incompatibility and its occurrence in nature. PLoS Genetics 10, e1004848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. 1949. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology 24, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W. 1909. Heredity and variation in modern lights. In: Seward AC, ed. Darwin and modern science. Cambridge: Cambridge University Press, 85–101. [Google Scholar]
- Bentham A, Burdett H, Anderson PA, Williams SJ, Kobe B. 2016. Animal NLRs provide structural insights into plant NLR function. Annals of Botany 119, 702–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Burdett H, Williams SJ, et al. . 2016. Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. The Plant Cell 28, 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D. 2007. Autoimmune response as a mechanism for a Dobzhansky–Muller-type incompatibility syndrome in plants. PLoS Biology 5, e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Cherkis K, Nishimura MT, Dangl JL. 2012. A new eye on NLR proteins: focused on clarity or diffused by complexity? Current Opinion in Immunology 24, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Bomblies K, Kim ST, et al. . 2014. Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159, 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Tran DT, Weigel D. 2016. Cooperation and conflict in the plant immune system. PLoS Pathogens 12, e1005452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Lin YS, et al. . 2014. A two-locus interaction causes interspecific hybrid weakness in rice. Nature Communications 5, 3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Shan JX, Zhu MZ, Shi M, Gao JP, Lin HX. 2013. Genetic and physiological analysis of a novel type of interspecific hybrid weakness in rice. Molecular Plant 6, 716–728. [DOI] [PubMed] [Google Scholar]
- Cohen SP, Liu H, Argueso CT, Pereira A, Vera Cruz C, Verdier V, Leach JE. 2017. RNA-seq analysis reveals insight into enhanced rice Xa7-mediated bacterial blight resistance at high temperature. PLoS ONE 12, e0187625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE. 2015. Effector-triggered immunity: from pathogen perception to robust defense. Annual Review of Plant Biology 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Danot O. 2015. How ‘arm-twisting’ by the inducer triggers activation of the MalT transcription factor, a typical signal transduction ATPase with numerous domains (STAND). Nucleic Acids Research 43, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. 1937. Genetics and the origin of species. New York: Columbia University Press. [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Eckardt N. 2017. Receptor-like kinases, ROS-RLK crosstalk, quantitative resistance, and the growth/defense tradeoff. The Plant Cell 29, 289–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi F, Chung E, Anderson RG, Li J, Wan L, Eitas TK, Gao Z, Dangl JL. 2017. Signaling from the plasma-membrane localized plant immune receptor RPM1 requires self-association of the full-length protein. Proceedings of the National Academy of Sciences, USA 114, E7385–E7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Gong H, Hu Y, et al. . 2017. Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Biology 18, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel DU. 1954. A new lethal combination in interspecific cotton hybrids. Genetics 39, 628–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CJ, Slootweg EJ, Goverse A, Baulcombe DC. 2013. Stepwise artificial evolution of a plant disease resistance gene. Proceedings of the National Academy of Sciences, USA 110, 21189–21194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano H, Mizuno N, Matsuda R, Shitsukawa N, Park P, Takumi S. 2012. Dysfunction of mitotic cell division at shoot apices triggered severe growth abortion in interspecific hybrids between tetraploid wheat and Aegilops tauschii. New Phytologist 194, 1143–1154. [DOI] [PubMed] [Google Scholar]
- Hellens R, Mullineaux P, Klee H. 2000. A guide to Agrobacterium binary Ti vectors. Trends in Plant Science 5, 446–451. [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. 2014. Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Molecular Plant 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S, Truong H, Danen HJ. 2007. Integrins: signaling, disease, and therapy. International Journal of Radiation Biology 83, 743–751. [DOI] [PubMed] [Google Scholar]
- Jalmi SK, Sinha AK. 2015. ROS mediated MAPK signaling in abiotic and biotic stress- striking similarities and differences. Frontiers in Plant Science 6, 769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins JG. 1953. Coastland—a new long staple cotton for the southeast. University of Georgia: Georgia Coastal Plain Experiment Station Bulletin 53. [Google Scholar]
- Jeuken MJ, Zhang NW, McHale LK, Pelgrom K, den Boer E, Lindhout P, Michelmore RW, Visser RG, Niks RE. 2009. Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. The Plant Cell 21, 3368–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhang TZ. 2003. Extraction of total RNA in cotton tissues with CTAB-acidic phenolic method. Cotton Science 15, 166–167. [Google Scholar]
- Kim KC, Fan B, Chen Z. 2006. Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiology 142, 1180–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knepper C, Savory EA, Day B. 2011. The role of NDR1 in pathogen perception and plant defense signaling. Plant Signaling & Behavior 6, 1114–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohel RJ, Richmond TR, Lewis CF. 1970. Texas marker-1. Description of a genetic standard for Gossypium hirsutum L. Crop Science 10, 670–671. [Google Scholar]
- Kourelis J, van der Hoorn RAL. 2018. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. The Plant Cell 30, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA. 1981. Genetics of D3 complementary lethality in Gossypium hirsutum and G. barbadense. Journal of Heredity 72, 299–300. [Google Scholar]
- Li X, Kapos P, Zhang Y. 2015. NLRs in plants. Current Opinion in Immunology 32, 114–121. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. 2002. Virus-induced gene silencing in tomato. The Plant Journal 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Ma JM. 2017. The fine mapping of two black shank resistance loci and identification of a hybrid lethality gene in tobacco. PhD thesis, North Carolina State University. [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, et al. . 2011. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host & Microbe 9, 187–199. [DOI] [PubMed] [Google Scholar]
- Muller HJ. 1942. Isolating mechanisms, evolution and temperature. Biology Symposium 6, 71–125. [Google Scholar]
- Niu CF, Wei W, Zhou QY, et al. . 2012. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant, Cell & Environment 35, 1156–1170. [DOI] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Foyer CH. 2016. Oxidative stress and antioxidative systems: recipes for successful data collection and interpretation. Plant, Cell & Environment 39, 1140–1160. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Brubaker CL, Wendel JF. 1993. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Molecular Biology Reporter 11, 122–127. [Google Scholar]
- Paterson AH, Wendel JF, Gundlach H, et al. . 2012. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. [DOI] [PubMed] [Google Scholar]
- Phillips LL, Merritt JF. 1972. Interspecific incompatibility in Gossypium. I. Stem histogenesis of G. hirsutum × G. gossypioides. American Journal of Botany 59, 203–208. [Google Scholar]
- Phillips LL, Reid RK. 1975. Interspecific incompatibility in Gossypium. II. Light and electron microscope studies of cell necrosis and tumorigenesis in hybrids of G. klotzschianum. American Journal of Botany 62, 790–796. [Google Scholar]
- Presgraves DC, Noor M. 2002. Patterns of postzygotic isolation in Lepidoptera. Evolution 56, 1168–1183. [DOI] [PubMed] [Google Scholar]
- Price TD, Bouvier MM. 2002. The evolution of F1 postzygotic incompatibilities in birds. Evolution 56, 2083–2089. [PubMed] [Google Scholar]
- Provine WB. 1991. Alfred Henry Sturtevant and crosses between Drosophila melanogaster and Drosophila simulans. Genetics 129, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Ye J, Geng YF, Sun YW, Gao SQ, Zhang BP, Chen W, Chua NH. 2012. Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiology 160, 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Blackman BK. 2010. Speciation genes in plants. Annals of Botany 106, 439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Willis JH. 2007. Plant speciation. Science 317, 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samora PJ, Stelly DM, Kohel RJ. 1994. Localization and mapping of the Le1 and Gl2 loci of cotton (Gossypium hirsutum L.). Journal of Heredity 85, 152–157. [Google Scholar]
- Sasa MM, Chippindale PT, Johnson NA. 1998. Patterns of postzygotic isolation in frogs. Evolution 52, 1811–1820. [DOI] [PubMed] [Google Scholar]
- Shao ZQ, Xue JY, Wu P, Zhang YM, Wu Y, Hang YY, Wang B, Chen JQ. 2016. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiology 170, 2095–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silow RA. 1941. The comparative genetics of Gossypium anomalum and the cultivated Asiatic cottons. Journal of Genetics 42, 259–358. [Google Scholar]
- Song L, Guo W, Zhang T. 2009. Interaction of novel Dobzhansky–Muller type genes for the induction of hybrid lethality between Gossypium hirsutum and G. barbadense cv. Coastland R4-4. Theoretical and Applied Genetics 119, 33–41. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X. 2012. How do plants achieve immunity? Defence without specialized immune cells. Nature Reviews. Immunology 12, 89–100. [DOI] [PubMed] [Google Scholar]
- Stelly DM. 1990. Localization of the Le2 locus of cotton (Gossypium hirsutum L.). Journal of Heredity 81, 193–197. [Google Scholar]
- Stephens SG. 1946. The genetics of ‘corky’. I. The New World alleles and their possible role as an interspecific isolating mechanism. Journal of Genetics 47, 150–161. [DOI] [PubMed] [Google Scholar]
- Stephens SG. 1976. Some observations on photoperiodism and the development of annual forms of domesticated cottons. Economic Botany 30, 409–418. [Google Scholar]
- Sukarta OCA, Slootweg EJ, Goverse A. 2016. Structure-informed insights for NLR functioning in plant immunity. Seminars in Cell & Developmental Biology 56, 134–149. [DOI] [PubMed] [Google Scholar]
- Świadek M, Proost S, Sieh D, et al. . 2017. Novel allelic variants in ACD6 cause hybrid necrosis in local collection of Arabidopsis thaliana. New Phytologist 213, 900–915. [DOI] [PubMed] [Google Scholar]
- Takahama U, Oniki T. 1997. A peroxidase/phenolics/ascorbate system can scavenge hydrogen peroxide in plant cells. Physiologia Plantarum 101, 845–852. [Google Scholar]
- Tran DTN, Chung EH, Habring-Müller A, Demar M, Schwab R, Dangl JL, Weigel D, Chae E. 2017. Activation of a plant NLR complex through heteromeric association with an autoimmune risk variant of another NLR. Current Biology 27, 1148–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang Z, Li F, et al. . 2012. The draft genome of a diploid cotton Gossypium raimondii. Nature Genetics 44, 1098–1103. [DOI] [PubMed] [Google Scholar]
- Waszczak C, Carmody M, Kangasjärvi J. 2018. Reactive oxygen species in plant signaling. Annual Review of Plant Biology 69, 209–236. [DOI] [PubMed] [Google Scholar]
- Yan H, Jia H, Chen X, Hao L, An H, Guo X. 2014. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant & Cell Physiology 55, 2060–2076. [DOI] [PubMed] [Google Scholar]
- Yi X, Du Z, Su Z. 2013. PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Research 41, W98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hu Y, Jiang W, et al. . 2015. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnology 33, 531–537. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dodds PN, Bernoux M. 2017. What do we know about NOD-like receptors in plant immunity? Annual Review of Phytopathology 55, 205–229. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cheng Y, Guo J, Yang E, Liu C, Zheng X, Deng K, Zhou J. 2014. Comparative transcriptome analysis to reveal genes involved in wheat hybrid necrosis. International Journal of Molecular Sciences 15, 23332–23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang Y, Fang B, Gannon P, Ding P, Li X, Zhang Y. 2010. Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRKY70. The Plant Cell 22, 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuellig MP, Sweigart AL. 2018. Gene duplicates cause hybrid lethality between sympatric species of Mimulus. PLoS Genetics 14, e1007130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.