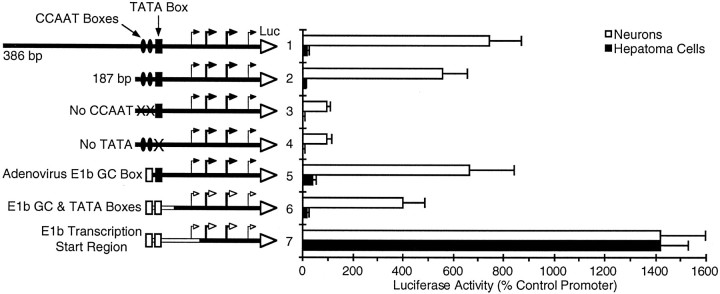
Fig. 2.
A 38 bp sequence located downstream of the GAP-43 TATA box can determine tissue-specific expression. The promoter constructs shown schematically were tested for their ability to drive the expression of a luciferase reporter gene (Luc) in neurons or hepatoma cells (same as Fig. 1). In construct3 both CCAAT consensus sequences have been mutated to C ACCT, and in construct 4 the TATA box sequence has been changed from TTTAAATATT to TT GCAA GCTT. Sequences borrowed from the adenovirus E1b promoter (constructs 5–7) are shown as open boxes or lines. The transcription start sites (bent arrows) for the rat 386 bp GAP-43 promoter are based on RNase protection analysis of human transcripts (Ortoft et al., 1993), because the rat and human promoters are highly homologous in this region. Heavier arrowsdesignate the more dominant transcription start sites. For the hybrid promoters with the adenovirus TATA box (constructs 6, 7), the open arrows are intended only as reference points, because we have not determined where transcription actually starts. The wild-type adenovirus E1b promoter normally initiates transcription from a single site 23 bp downstream of its TATA box (Wu et al., 1987).