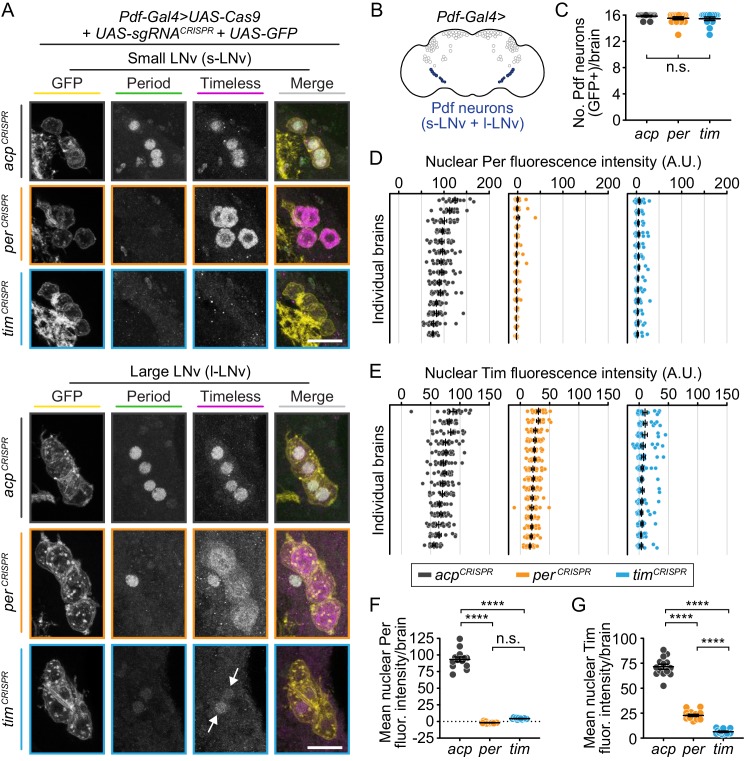
Figure 5. CRISPR-mediated disruption of per or tim in Pdf+ neurons leads to efficient loss of the targeted protein.
(A) Maximum intensity projections of Pdf-Gal4-driven GFP+ small and large ventral lateral neurons (s- and l-LNv) with immunohistochemistry for GFP (yellow), Period (green) and Timeless (magenta) at ZT0. Scale bar = 10 μm; arrows indicate CRISPR-targeted nuclei with residual protein signal. (B) Diagram showing Pdf-Gal4 expression in the 4 large and four small ventral lateral neurons (l- and s-LNv), bilaterally. (C) Quantification of the number of GFP+ neurons per brain (acp n = 14; per n = 15; tim n = 13 brains). (D and E) Background-subtracted nuclear fluorescence intensity of Per (D) or Tim (E) at ZT0 in GFP+ neurons, grouped by brain with mean ± SEM shown. (F and G) Mean nuclear fluorescence intensity of Per (G) or Tim (H) at ZT0 in GFP+ neurons, averaged per brain (acp n = 14; per n = 16; tim n = 14 brains). ****: p<0.0001; n.s.: not significant, p>0.05. Significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test; reported p-values are multiplicity adjusted to account for multiple comparisons.