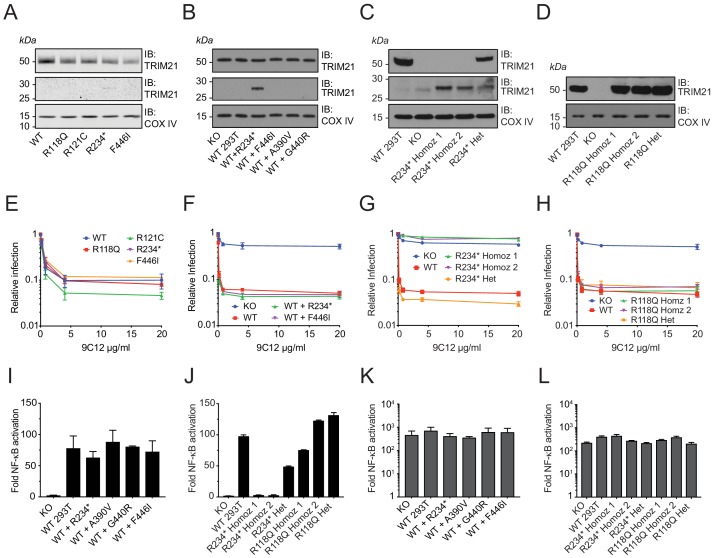
Figure 9. Natural missense variants do not exert dominant negative effect.
(A–D) Immunoblot (IB) for TRIM21 and COX IV (loading control) in (A) selected LCLs (B) Transduced WT 293T cell lines stably expressing TRIM21 variants under the native TRIM21 promoter. (C–D) CRISPR gene-edited 293T clones expressing the R234* and R118Q variant respectively. (E–H) AdV neutralization in the presence of anti-adenovirus hexon monoclonal IgG 9C12 in (E) selected LCLs. (F) WT 293T cells expressing the R234* and F446I variant. (G–H) CRISPR gene-edited 293T clones. Relative infection levels quantified by flow cytometry and normalized to virus only condition. Data compiled from at least two independent experiments (mean ± SEM). (I–L) Activation of NF-κB signaling by 9C12 coated AdV5 (I–J) or human TNF-α (K–L) in the respective cell lines. Data compiled from two independent experiments (mean ± SEM). Data provided in Figure 9—source data 1.