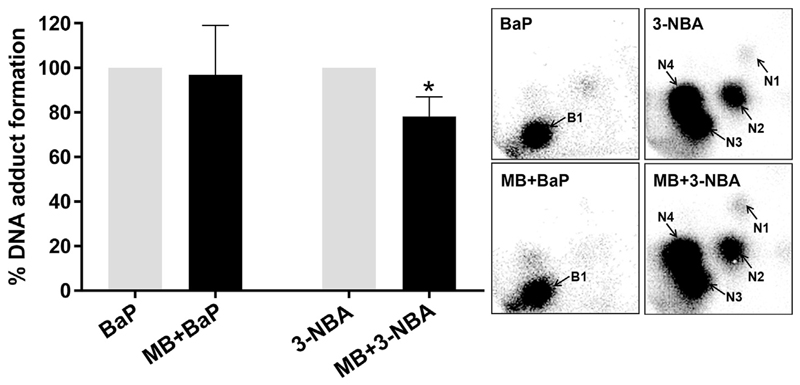
Fig. 5.
Effect of PS-MB (MB; 50 μg mL−1) on BaP- and 3-NBA-derived DNA adduct formation (%) in fish intestinal RTgutGC cells exposed for 48 h 32P-postlabelling was used to assess DNA adduct formation; for BaP-derived DNA adducts nuclease P1 digestion, for 3-NBA-derived DNA adducts butanol extraction was used as enrichment procedure. Values represent mean ± SD (n = 4) derived from four independent experiments with cells from different passage numbers. For statistical analysis the adduct data was normalised to 1.0, data then log2 transformed and analysed using a single sample t-test with Bonferroni correction against the population control mean of 0 (*p < 0.05, different from cells treated with 3-NBA alone). Inserts: Autoradiographic profiles of DNA adducts formed in fish gill RTgill-W1 and intestinal RTgutGC cells after exposure; the origin, at the bottom left-hand corner, was cut off before exposure. B1, 10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydro-BaP (dG-N2-BPDE); N1, 2′(2′-deoxyadenosine-N6-yl)-3-aminobenzanthrone (dA-N6-3-ABA); N2, as-yet unidentified adenine adduct derived from nitroreduction; N3, N-(2′-deoxyguanosine-N2-yl)-3-aminobenzanthrone (dG-N2-3-ABA); N4, N-(2′-deoxyguanosin-8-yl)-3-aminobenzanthrone (dG-C8-N-3-ABA).