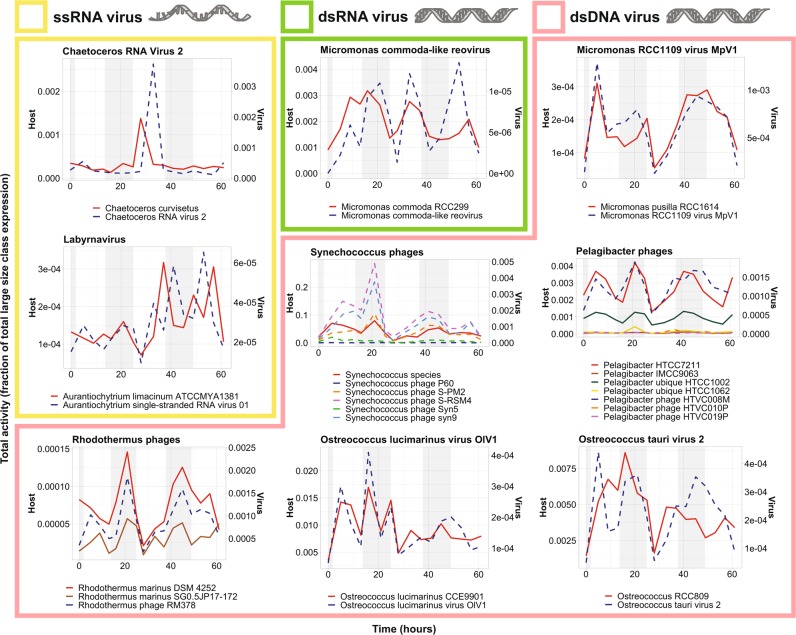
Fig. 6.
Virus/host dynamics in the large size class. Viruses and hosts are annotated as the closest reference available in our database, as determined by LPI. Library normalized expression of ORFs classified as ssRNA (yellow), dsRNA (green), and dsDNA (pink) viruses and their putative hosts by LPI are shown. Putative host expression is represented by solid lines and corresponds to left y-axes; virus expression is represented by dashed lines and corresponds to the right y-axes. Night hours are shaded in gray. Note that OtV2 infects RCC393, an Ostreococcus Clade OII species, not O. tauri. OtV2 was isolated against Ostreococcus Clade OII isolate RCC393 [94] which has 99% 18S rDNA identity to the genome sequenced Clade OII isolate RCC809 used in our mapping analysis. Likewise, the M. commoda-like reovirus infects the strain LAC38, which was initially misreported as being M. pusilla and has been renamed here according to proper species assignment of the host [87]. Interestingly, while picoprasinophyte populations mapping most closely to Micromonas pusilla were coactive with a dsDNA virus, populations mapping most closely to Micromonas commoda (RCC299) were coactive with a dsRNA virus [87, 95]