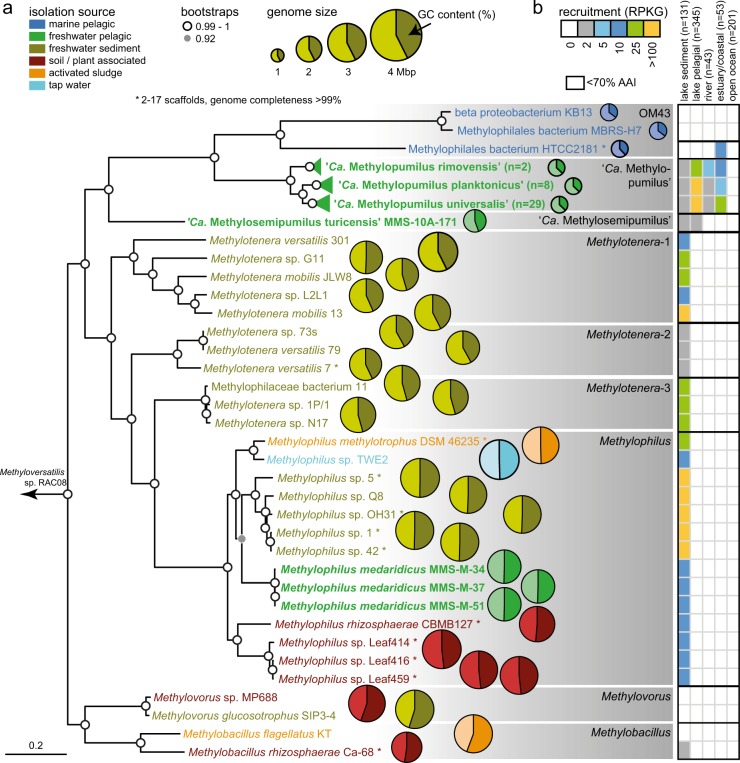
Fig. 1.
Phylogeny of Methylophilaceae and their occurrence in different environments. a Phylogenomic tree based on 878 common concatenated proteins (351,312 amino acid sites) with Methyloversatilis sp. RAC08 as outgroup. The 39 complete genomes of “Ca. Methylopumilus sp.” are collapsed to the species level, see Fig. S1 for a complete tree. Different genera (70% AAI cutoff, Fig. S2) are marked by gray boxes. Isolation sources of strains are indicated by different colors and incomplete genomes consisting of <17 contigs (estimated completeness >99%) are marked with asterisks. Bootstrap values (100 repetitions) are indicated at the nodes, the scale bar at the bottom indicates 20% sequence divergence. The genome sizes for all strains are shown with circles of proportional size and GC content is depicted within each circle. b Fragment recruitment of public metagenomes from freshwater sediments (n = 131), lake pelagial (n = 345), rivers (n = 43), estuaries and coastal oceans (n = 53), and open oceans (n = 201). Maximum RPKG values (number of reads recruited per kb of genome per Gb of metagenome) for each ecosystem are shown for each genome