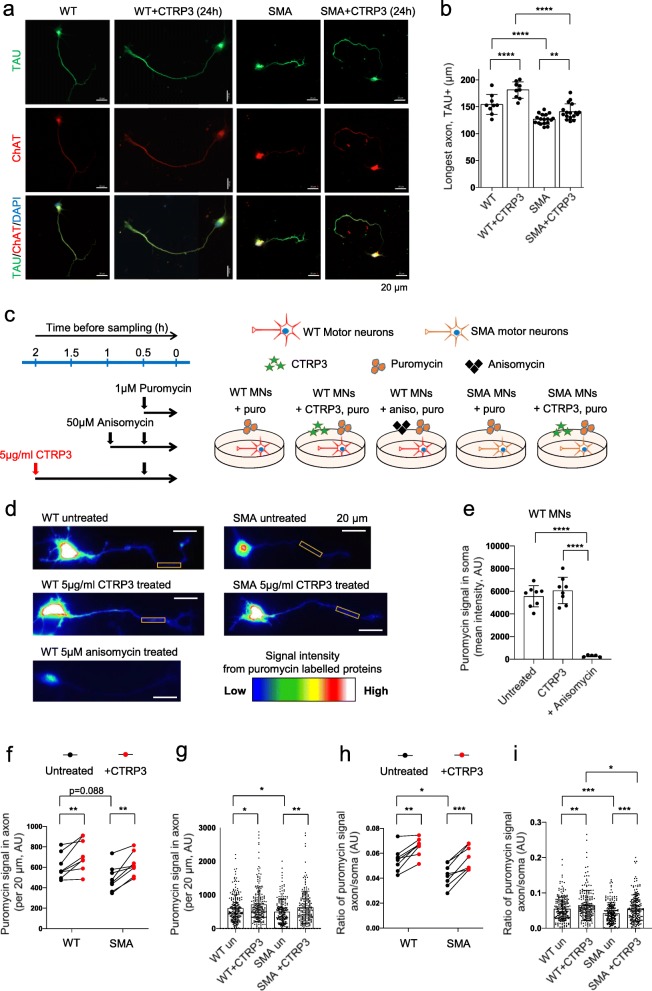
Fig. 6.
CTRP3 stimulates axonal growth and protein synthesis of motor neurons. a Axon outgrowth assay with WT and SMA motor neurons treated with 5 μg/ml hCTRP3 for 24 h (2DIV-3DIV). Representative images of motor neurons stained with anti-Tau (green), anti-ChAT (red) and DAPI (blue). Scale bars: 20 μm. b Dot plot bar graph summarizes axon outgrowth analysis. Each dot represents the average axon length of 20–25 neurons in a coverslip. Bar graphs depict the mean ± s.d. (WT: n = 9, N = 3; SMA: n = 17, N = 4; n = number of coverslips, N = number of independent cultures). Two way ANOVA with Sidak’s multiple comparison test was used to determine statistical significance; **p < 0.01, ****p < 0.0001. c Scheme of SUnSET assay to investigate the effect of CTRP3 on axonal protein synthesis. Motor neurons (3DIV) were treated with 5 μg/ml hCTRP3 for 2 h and newly synthesized proteins were labelled with 1 μM puromycin for 30 min before analysis. Data are compared with non-CTRP3 treated cells. As a negative control, protein synthesis was blocked with 50 μM anisomycin for 1 h before analysis. d Representative images of WT and SMA motor neurons immunologically stained with anti-puromycin antibody. Rainbow scale heatmap indicating the intensity of SUnSET signals. e Dot plot bar graph represents the mean intensity of the SUnSET signal in soma of WT motor neurons. Each dot represents the average value of 24–28 neurons from an independent culture and bar graphs depict the mean ± s.d. (Untreated n = 8/N = 8, CTRP3 treated n = 8/N = 8, anisomycin treated n = 5/N = 5, n = number of coverslips, N = number of independent cultures, 25 neurons per coverslip). Ordinary one-way ANOVA and Uncorrected Fisher’s LSD were used to determine statistical significance. ****p < 0.0001, f-g Quantification of the mean intensity of the SUnSET signal in axons (per 20 μm). f SUnSET signal in axon (WT N = 8, SMA N = 8, N = number of independent cultures). Each dot represents average data from one independent neuron culture. Therefore, data were only compared in the same culture with and without CTRP3 treatment. Two-tailed paired student’s t-test was used to determine statistical significance, **p < 0.01. To compare WT and SMA, unpaired t-test was used to determine statistical significance, and it was not significant. g Each dot represents a value from a neuron, WT untreated n = 204, WT CTRP3 treated n = 293, SMA untreated n = 200, SMA CTRP3 treated n = 200. Two way ANOVA with Sidak’s multiple comparison test was used to determine statistical significance; *p < 0.05, **p < 0.01, h-i Quantification of the ratio of the SUnSET signal in axons (per 20 μm) compared to average signal in soma. h Ratio between soma and axons were increased by CTRP3 treatment, WT N = 8, SMA N = 8. Each dot represents average data from one independent neuron culture. Therefore, data were only compared in the same culture with and without CTRP3 treatment. Two-tailed paired student’s t-test was used to determine statistical significance. ***p < 0.001 and **p < 0.01. To compare WT and SMA, unpaired t-test was used to determine statistical significance. *p < 0.05. i Ratio between soma and axons in individual neurons, each dot represents a value from a neuron, WT untreated n = 204, WT CTRP3 treated n = 293, SMA untreated n = 200, SMA CTRP3 treated n = 200. Two way ANOVA with Sidak’s multiple comparison test was used to determine statistical significance; *p < 0.05, **p < 0.01, ***p < 0.001