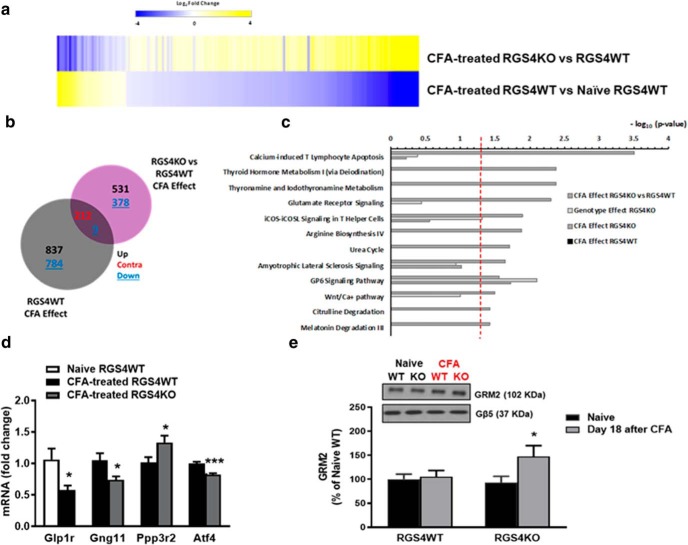
Figure 6.
RGS4 in the THL modulates gene expression patterns of peripheral inflammation. a, b, Heat map analysis (a) and Venn diagrams (b) showing the effect of peripheral inflammation in RGS4WT and RGS4KO THL (cutoff p value <0.05; cutoff log2fold <−0.25 and >0.25). The overall patterns of gene expression regulation were distinct between RGS4KO and RGS4WT groups. Interestingly, the commonly regulated genes are contraregulated by peripheral inflammation between genotypes (212 contraregulated genes). c, IPA of the set of genes affected by CFA in the RGS4KO group compared with RGS4WT predicts pathways that are selectively modulated upon RGS4 gene ablation in the RGS4KO group (cutoff: p value <0.003). d, qPCR validation of a subset of genes in a separate cohort of male animals (naive RGS4WT and CFA-treated RGS4WT: Glp1r (unpaired t test: t(11) = 2.735, *p = 0.02); CFA-treated RGS4WT and CFA-treated RGS4KO (unpaired t test: Gng11 t(24) = 2.486, *p = 0.02; Ppp3r2 t(10) = 2.364, *p = 0.04; Atf4 t(9) = 5.236, ***p < 0.001; n = 5–13/group. e, Western blot analysis in THL synaptosomes from naive and CFA groups of RGS4WT and RGS4KO mice reveals an upregulation of mGluR2 at 18 d after the induction of peripheral inflammation in the RGS4KO group. Two-way ANOVA followed by Bonferroni post hoc tests: F(1,59) = 3.505, *p = 0.04, n = 15–16/group.