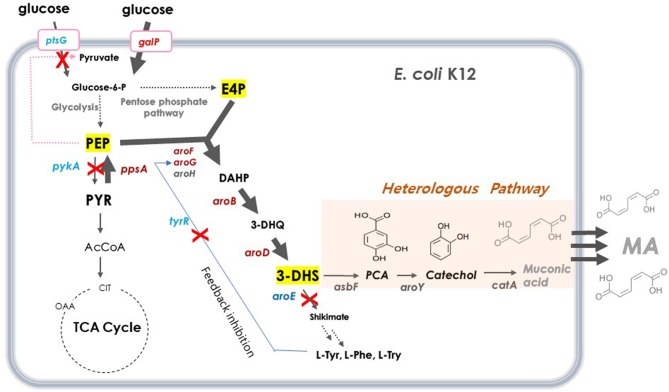
Figure 1.
Schematic overview of metabolic pathway for DHS biosynthesis in E. coli. Red crosses denote disrupted genes and bold blue arrows denote steps that are overexpressed. Dashed arrows represent two or more steps. G6P, glucose-6-phosphate; PEP, phosphoenolpyruvate; PYR, pyruvate; AcCoA, acetyl-coenzyme A; OAA, oxaloacetate; CIT, citrate; E4P, erythrose4-phosphate; DAHP, 3-deoxy-D-arabinoheptulosonate-7-phosphate; DHQ, 3-de-hydroquinate; DHS, 3-dehydroshikimate; SA, shikimic acid; PCA, protocatechuate; CA, catechol, MA, muconic acid. ptsG, glucose specific sugar: phosphoenolpyruvate phosphotransferase; galP, D-galactose transporter; zwf, glucose-6-phosphate 1-dehydrogenase; tktA, transketolase; tyrR, tyrosine dependent transcriptional regulator; pykF, pyruvate kinase 1; pykA, pyruvate kinase 2; ppsA, phosphoenolpyruvate synthase; pckA, phosphoenolpyruvate carboxykinase; aroG, DAHP synthase; aroF, DAHP synthase; aroB, DHQ synthase; aroD, DHQ dehydratase; aroE, shikimate dehydrogenase: asbFEopt, 3-dehydroshikimate(DHS) dehydratase; pcaG/H, codon-optimized PCA deoxygenase; aroYEopt, codon-optimized PCA decarboxylase; catAEopt, codon-optimized CA 1,2 -dioxygenase.