FIGURE 1.
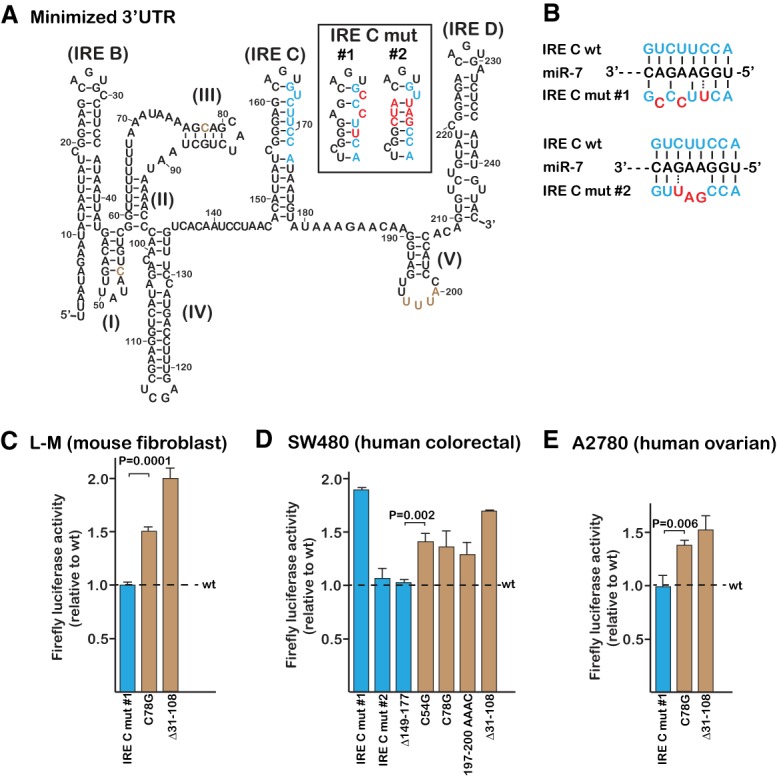
The proposed miR-7-5p binding site on the TfR1 mRNA is not functional. (A) Secondary structure of the minimized TfR1 mRNA previously determined to be required for efficient iron-responsiveness (Casey et al. 1989). Brown, sites of point mutations to the three 5 bp stem–loops (I, III, and V) that were previously identified as essential for iron-responsive degradation in mouse L-M cells (Rupani and Connell 2016); blue, the proposed miR-7-5p binding site. The RNA structure and labeling are based on earlier established convention (Horowitz and Harford 1992; Schlegl et al. 1997). Inset shows the predicted secondary structure of IRE C resulting from the mutations (red) used to disrupt the proposed miR-7-5p binding site; IRE C mut #1 is the Miyazawa et al. mutation. (B) Predicted base-pairing between the miR-7-5p seed sequence and the proposed TfR1 mRNA binding site should be disrupted by both IRE C mut #1 and #2. (C) Mutation of the proposed miR-7-5p binding site (blue) has no significant impact on mRNA stabilization when assayed in mouse L-M cells. Representative earlier mutations that do not overlap with the proposed binding site but impede TfR1 mRNA degradation were assayed in parallel (brown). (D) Although IRE C mut #1 appears to stabilize the reporter RNA in SW480 cells, neither IRE C mut #2 nor the complete deletion of the binding site (Δ149–177) has a significant effect. (E) IRE C mut #1 has no effect when assayed within A2780 cells. All error bars represent ±SEM of three biological replicates.
