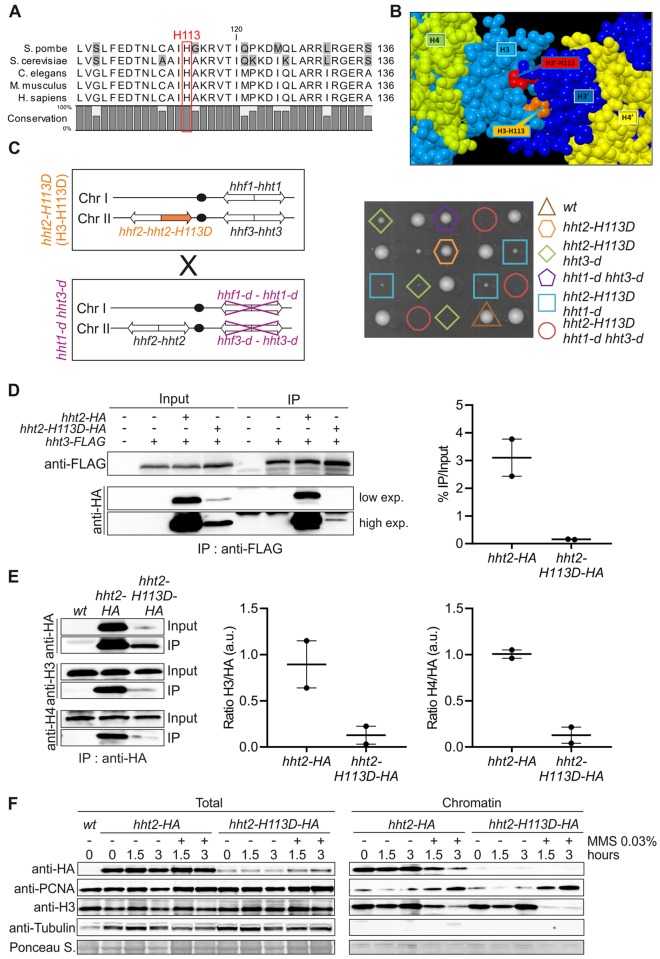
Fig 2. The mutated histone H3-H113D destabilizes (H3-H4)2 tetramer.
(A) Alignment of yeast, worm, mouse and human histone H3. Red rectangle indicates the position of histidine 113. UniProtKB access codes used are S. pombe: P09988; S. cerevisiae: P61830; C.elegans: P08898; M. musculus: P84228 and H. sapiens: Q71DI3. (B) View of the H3:H3’ interface. The interface between the two histones H3, colored here in light (H3) and dark blue (H3’), maintains H3 and H4 in the tetramer form (H3-H4)2. In the H3:H3’ interface, H113 of each histone (orange for H113 in H3 and red for H113 in H3’) is deeply buried in the adjacent histone partner. For clarity, the other histones H2A and H2B, as well as DNA, are not represented. (C) Left panel: schematic of the strains crossed. Right panel: spore viability analysis of indicated genotypes. (D) Left panel: association of H3-FLAG with H3-HA and H3-H113D-HA in indicated strains. Right panel: quantification expressed in arbitrary unit (a.u.). Individual values are plotted ± the range. (E) Left panel: association of H3-HA and H3-H113D-HA with untagged H3 and H4 in indicated strains. Right panels: quantification expressed in arbitrary unit (a.u.). Individual values are plotted ± the range. (F) Chromatin association of analyzed proteins in indicated strains and conditions (hours upon MMS addition or not). Two independent biological replicates were performed and representative blots are shown. See S2 Fig for structural impact of H3-H113D.