Fig. 2. Sequence alignment of P-Rex1 with its close homologs along with other annotated characteristics.
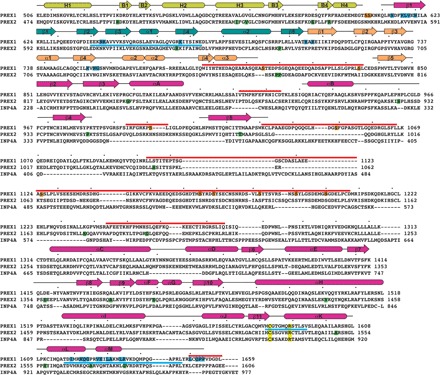
Although residues 38 to 505 of P-Rex1 were present in the protein used for cryo-EM analysis, they were not observed in the high-resolution reconstruction and are not shown here. Clustal Omega was used to align the sequences of human P-Rex1 (UniProtKB ID Q8TCU6), P-Rex2 (ID Q70Z35), and inositol 3,4-bisphosphate 4-phosphatase (INPP4A; ID Q96PE3). Residues 1 to 227 of INPP4A, corresponding to its N-terminal C2 domain, were excluded. The dots above the alignment correspond to every 10th amino acid in the P-Rex1 sequence. The secondary structure elements observed in P-Rex1 are shown above the alignment, with α helices depicted as rounded rectangles, β strands as arrows, and coil as a black line. They are colored by their corresponding domain as defined in Fig. 1. The absence of indicated secondary structure indicates that these residues were not observed in the structure. Thick red bars above the sequence correspond to P-Rex1 regions that are >90% exchanged with deuterium after 1000 s (see fig. S9 and data file S1). Thick blue bars indicate regions that are significantly stabilized (4% or greater protection at 1000 s) during HDX-MS in the presence of Gβγ (see Fig. 5 and data files S1 and S2). Residues highlighted in blue correspond to those that bury ≥5 Å2 of accessible surface area in the Gβγ complex (out of a total of 1000 Å2 buried accessible surface area on P-Rex1). Most also correspond to regions that are stabilized in HDX-MS upon complex formation. Residues highlighted in yellow correspond to canonical cysteine and arginine active site residues found in PTEN and Legionella phosphoinositide phosphatases SidF and SidP. Residues in P-Rex1 reported to be phosphorylated are highlighted in orange and are found in the more dynamic loops of the structure where protein kinases would have easier access. P-Rex2 residues that are associated with mutation in cancer patients are highlighted in green. Gβγ-binding residues are not conserved in INPP4A, and its phosphatase active site is much more basic than that of P-Rex1 in part due to the presence of two lysines in its P loop, analogous to those conserved in PTEN, SidF, and SidP, consistent with their robust phosphatase activity.
