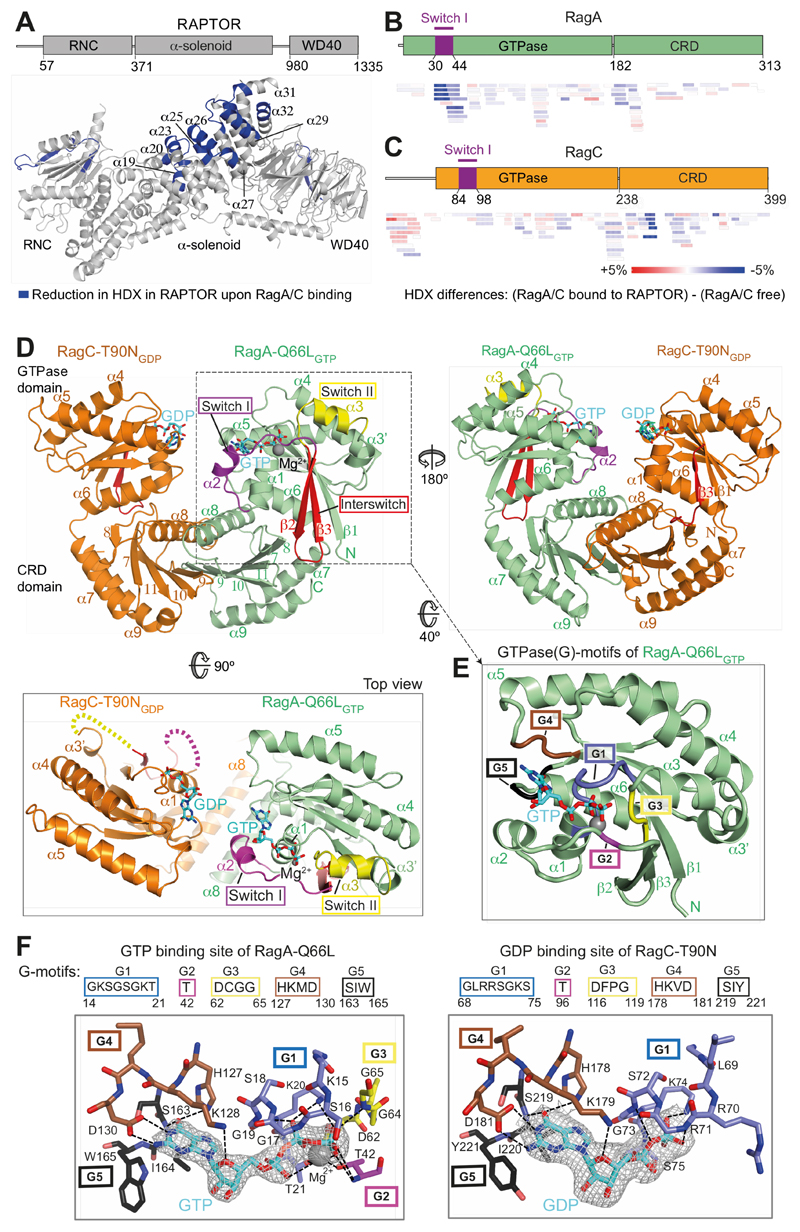
Fig. 1. The crystal structure of a RagA/C heterodimer and HDX-MS analysis of its interaction with RAPTOR.
(A) HDX-MS identified regions protected from HDX in the RAPTOR/RagA-Q66LGTP/RagCT90NGDP complex. Decreases in HDX (blue) of RAPTOR upon RagA/C binding are depicted on the RAPTOR structure (from PDB ID: 6BCX).
(B, C) Differences in HDX for RagA-Q66LGTP (B) and RagC-T90NGDP (C) upon RAPTOR binding for all the peptides at 0.3 s in D2O. Decreases in HDX are depicted in shades of blue, and increases are in shades of red.
(D) The crystal structure of RagA-Q66LGTP/RagC(34-399)-T90NGDP, highlighting the ordered switches in RagAGTP compared with disordered switches in RagC-T90NGDP oncogenic mutant.
(E) The conserved G-motifs of RagA-Q66LGTP that make up the nucleotide binding pocket.
(F) The 2mFo-DFc map (contoured at 1.2σ) for the GTP of RagA and the GDP of RagC together with the putative H-bonds that they make to the G-motifs. These are much more extensive for GTP than for GDP.