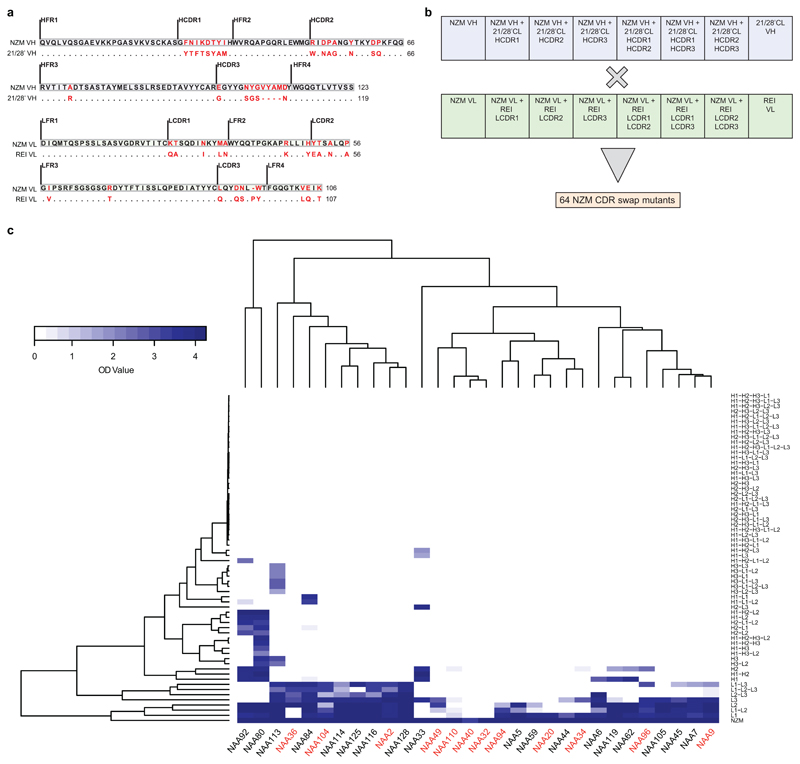
Extended Data Fig. 1. Epitope mapping of NZM-specific antibodies.
a, Alignment of NZM heavy and light chain variable regions (NZM VH and NZM VL) to the human scaffold antibody counterparts (21/28’CL and REI) used for NZM humanization. Mutated residues are shown in red. Dots indicate the same residue. b, Scheme of the 8 heavy and 8 light chains variants of NZM that were combined in an 8x8 matrix to express 64 different NZM CDR swap variants. c, Cluster analysis of binding of 30 antibodies isolated from patient A to the 64 NZM swap variants by ELISA. BAbs and NAbs are indicated on the x-axis in red and black, respectively. The NZM swap variants are shown on the right y-axis (H, heavy chain; L, light chain; 1, CDR1; 2, CDR2; 3, CDR3). Optical density (OD) values are shown with a two-color gradation scale from minimum (white) to maximum (blue).