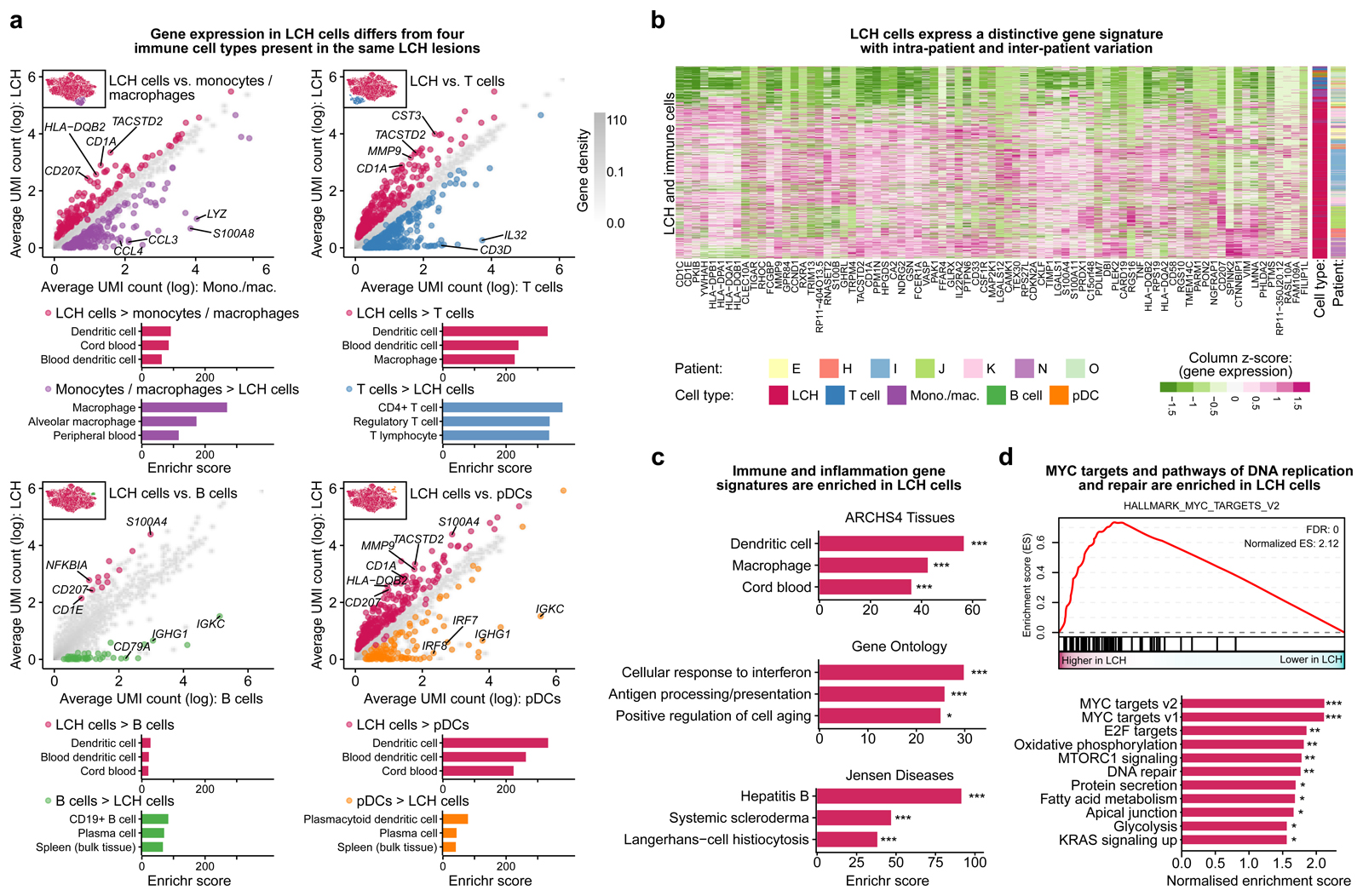
Figure 2. Characteristic gene expression patterns distinguish LCH cells from other immune cells present in LCH lesions.
A) Scatterplots comparing gene expression (mean normalized UMI counts plotted on logarithmic scale) between LCH cells and non-LCH immune cell populations. Differentially expressed genes specific for LCH cells are colored in red, whereas genes specific for the immune cell populations are colored in violet (monocyte/macrophages), blue (T cells), green (B cells), and orange (plasmacytoid dendritic cells). The top three enriched categories from the ARCHS4 tissue database based on Enrichr are shown below.
B) Heatmap showing the expression of the genes in the LCH gene signature (genes overexpressed in LCH cells compared to at least three of the four immune cell populations in panel A) in the LCH and immune cells analyzed (rows). Cell type and patient are indicated by the colored bars on the right.
C) Enrichment analysis with Enrichr for the LCH signature genes using three databases (top to bottom: ARCHS4 Tissues, Gene Ontology, or Jensen Diseases), ordered by the Enrichr combined score58. FDR-adjusted p-value: *, p < 0.1; ***, p < 0.001. Further details are provided in Supplementary Table S3.
D) Gene set enrichment analysis (GSEA) for LCH cell gene expression compared to immune cells using MSigDB hallmark signatures, including a GSEA plot for the most significant gene set (MYC_TARGETS_V2, top) and all significant enrichments visualized in the bar plot (bottom). FDR-adjusted p-value: *, p < 0.1; **, p < 0.05; ***, p < 0.001