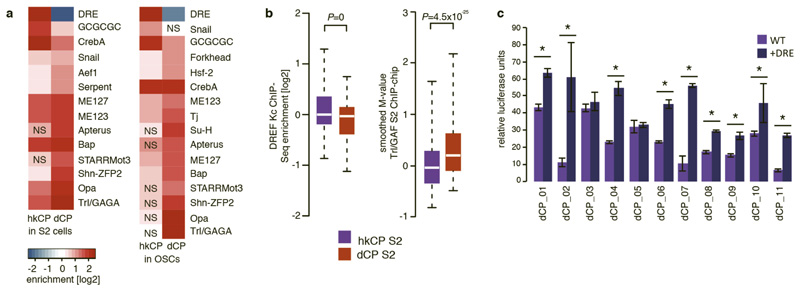
Extended Data Figure 10. The activity of hkCP and dCP enhancers are dependent on DRE and Trl/GAGA motifs, respectively.
a, Differential motif enrichment in distally located hkCP and dCP-specific enhancers (as Fig. 5a but assessing enrichments of the same motif PWMs exclusively at distal enhancers >500bp away from the closest TSSs). Key motifs including DRE and Trl/GAGA are also differentially enriched in distal hkCP and dCP-specific enhancers (NS: non-significant [FDR-corrected hypergeometric P>0.01]; S2 cells: hkCP n=790, dCP n=3013; OSCs: hkCP n=556, dCP n=2555). b, Distal hkCP and dCP-specific enhancers are differentially bound by DREF and Trl/GAF, respectively. ChIP enrichments of DREF (left) and Trl/GAF (right) at S2 hkCP and dCP-specific enhancers that are distal (>500bp) from the closest TSSs. Equivalent to Figure 5b, but considering exclusively TSS-distal enhancers to exclude potentially confounding effects for TSS-proximal enhancers for which it is not possible to discern whether binding occurs due to the enhancer sequence or core promoter function. The differential binding between DREF and Trl/GAF to hkCP and dCP-specific enhancers respectively is also found in Kc cells, in which the DREF ChIP-seq experiment had been performed (data not shown). c, Addition of DRE motifs to dCP enhancers increases their activity towards hkCP. Relative luciferase activity values (Firefly/Renilla [FF/RL]) for 11 dCP enhancers without DRE motifs (WT, light purple) and with 3 DRE motifs flanking the enhancers on each side (+DRE, dark purple). Asterisks (*) indicate statistical significance (P<0.05 via one-sided unpaired Student’s t-test); error bars denote the s.d. of three biological replicates.