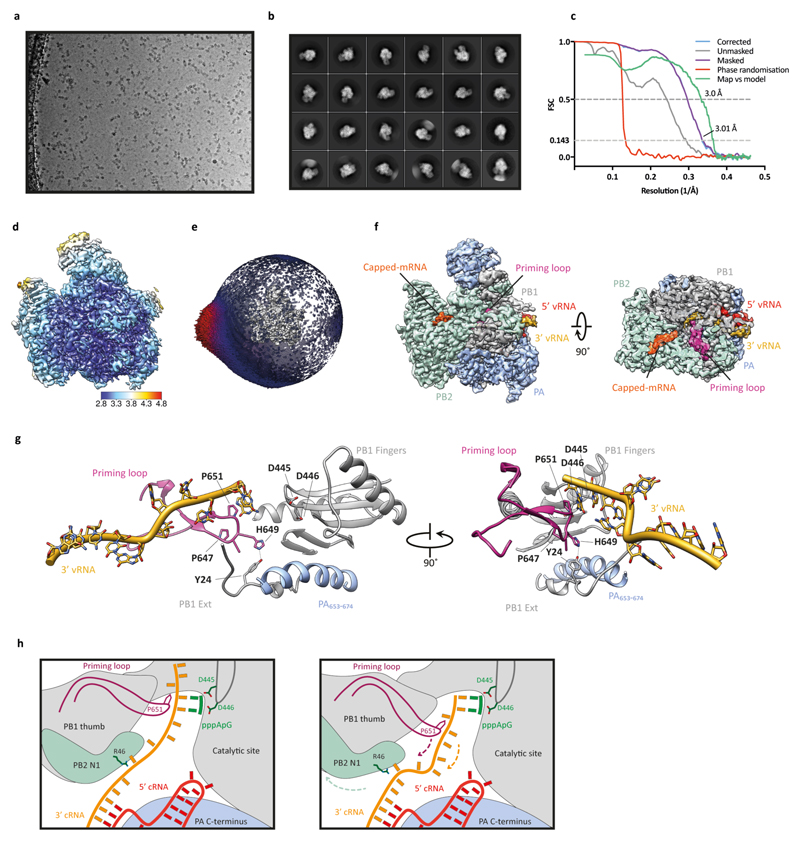
Extended Data Fig. 7. Single-particle cryo-EM analysis of human H3N2 FluPolA bound to vRNA promoter.
a, Representative micrograph of vRNA-bound FluPolA heterotrimer particles embedded in vitreous ice. b, Representative 2D class averages. c, FSC curves for 3D reconstruction using gold-standard refinement in RELION, indicating overall map resolution of 3.01 Å and the model-to-map FSC. Curves are shown for phase randomisation, unmasked, masked and phase-randomisation-corrected masked maps. d, 3D reconstruction locally filtered and coloured according to RELION local resolution. e, Angular distribution of particle projections with the cryo-EM map shown in grey. f, Cryo-EM map of vRNA-bound FluPolA heterotrimer revealing the presence of a fully resolved priming loop. g, Close-up views highlighting the stacking of the 3′ vRNA by the priming loop. h, Cartoon illustration of the role of polymerase dimerisation in template realignment during replication initiation on a cRNA template. Base-pairing between the 5′ and 3′ cRNA positions bases 4 and 5 of the 3′ cRNA next to the catalytic aspartates (PB1 amino acid residues D445 and D446) in the active site to allow internal replication initiation by the synthesis of a pppApG dinucleotide. The priming loop stacks the cRNA template through PB1 amino acid P651 (left panel). Rotation of the PB1 thumb/PB2-N1 domain triggered by polymerase dimerisation results in a movement of the priming loop and backtracking of the stacked template (arrows). Backtracking is also facilitated by an interaction of PB2 amino acid residue R46 with the 3′ cRNA introducing a ‘kink’ in the template. Backtracking positions bases 1 and 2 of the cRNA template opposite the pppApG dinucleotide that remains coordinated by the catalytic aspartates. The resulting replication complex is ready to extend the pppApG dinucleotide by incorporating the next incoming NTP (right panel).