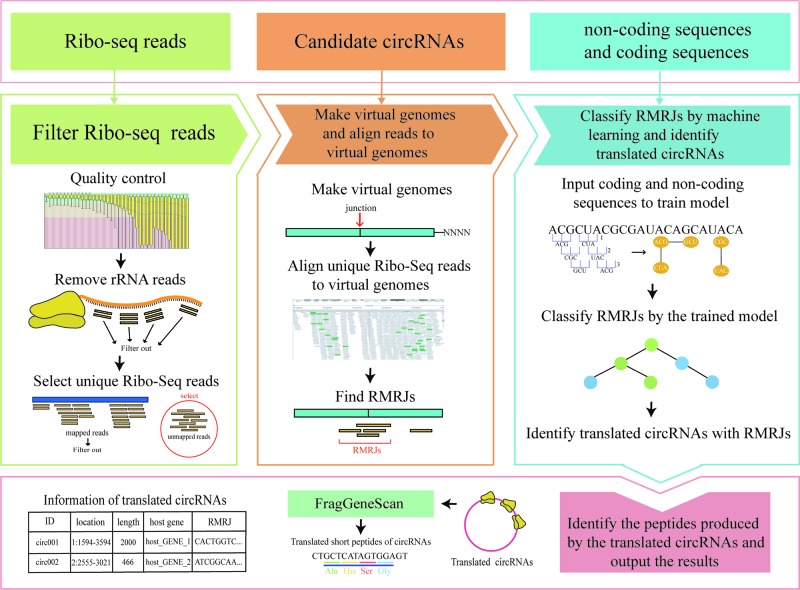
Figure 1.
The workflow of CircCode. The top layer represents the input file required for each step of CircCode. The middle layer is divided into three parts, and each part represents a different stage of operation. From left to right, the first part represents the filtering of the Ribo-seq data; the quality control is executed by Trimmomatic, and the rRNA reads are removed by bowtie. The second part represents the steps used to produce the virtual genome and align the filtered reads to the virtual genome with STAR. The last part represents the identification of translated circRNAs by machine learning. The bottom layer represents the last step used to predict the peptides translated from the circRNAs and the final output results, including information on translated circRNAs and their translation products.