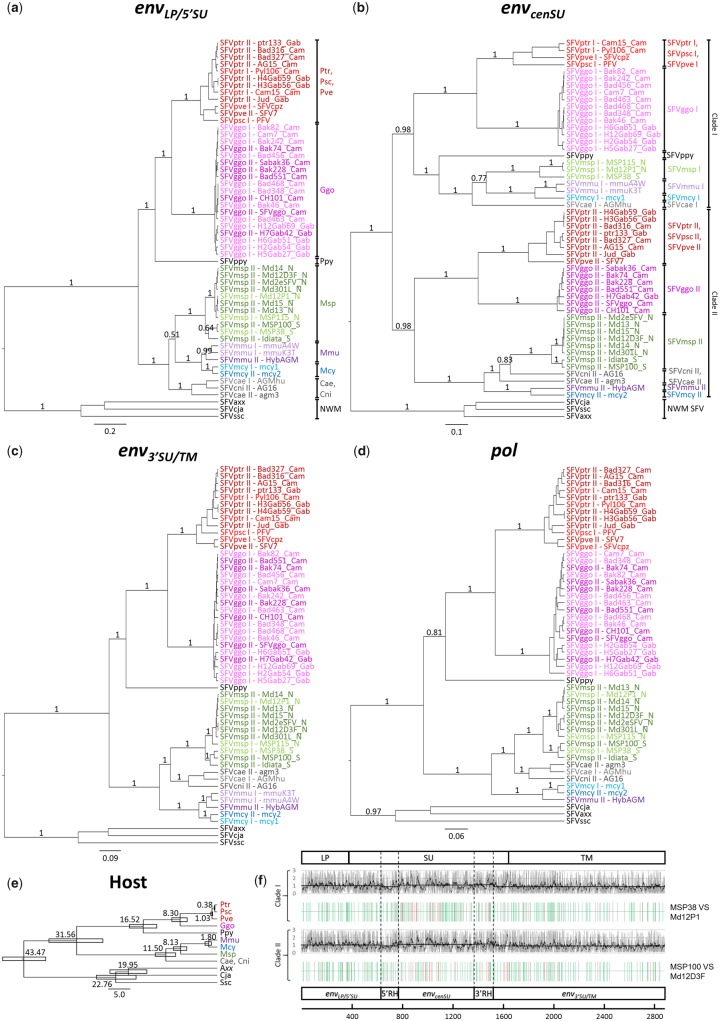
Figure 2.
env and pol phylogenies. Bayesian phylogenies were estimated from (a) the envLP/5′SU alignment, (b) the envcenSU alignment, (c) the env3′SU/TM alignment, and (d) the pol alignment, which were 630, 600, 1,362, and 425 nt long, respectively. Numbers on branches are Bayesian posterior probability clade support values. Scale bars are in units of substitutions per site. For envLP/5′SU, env3′SU/TM, and pol phylogenies, sequences segregate according to their host species, forming monophyletic clades. In envcenSU phylogeny, OWMA FVs form two well-supported distinct clades, clades I and II. In addition to the label ‘I’ and ‘II’ in their names, light and darker colours indicate if sequences belong to clade I or II, respectively. (e) Host phylogeny. The topology of the host tree and the divergence dates were estimated elsewhere (see the references in Supplementary Table S2). (f) Site-wise evolutionary rate estimates of the env gene for variant I (top), and variant II (bottom). See legend to Fig. 1. The figure also shows the distributions of silent (green) and non-silent (red) substitutions, computed by using Highlighter (https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html): top, comparison between clade I SFVmsp MSP38 and SFVmsp Md12P1; bottom, between clade II SFVmsp MSP100 and SFVmsp Md12D3F. The distributions are nearly uniform. SFV, simian foamy virus; Psc, Pan troglodytes schweinfurthii chimpanzee (red); Pve, Pan troglodytes verus chimpanzee (red); Ptr, Pan troglodytes troglodytes chimpanzee (red); Ggo, Gorilla gorilla gorilla gorilla (pink); Ppy, Pongo pygmaeus orangutan (black); Msp, Mandrillus sphinx mandrill (green); Mmu, Macaca mulatta macaque (purple); Mcy, Macaca cyclopis macaque (blue); Cae, Chlorocebus aethiops Grivet (grey); Cni, Cercopithecus nictitans Greater spot-nosed monkey (grey); NWM, New World Monkey (black); Axx, Ateles spider monkey (black); Cja, Callithrix jacchus marmoset (black); Scc, Saimiri sciureus squirrel monkey (black).