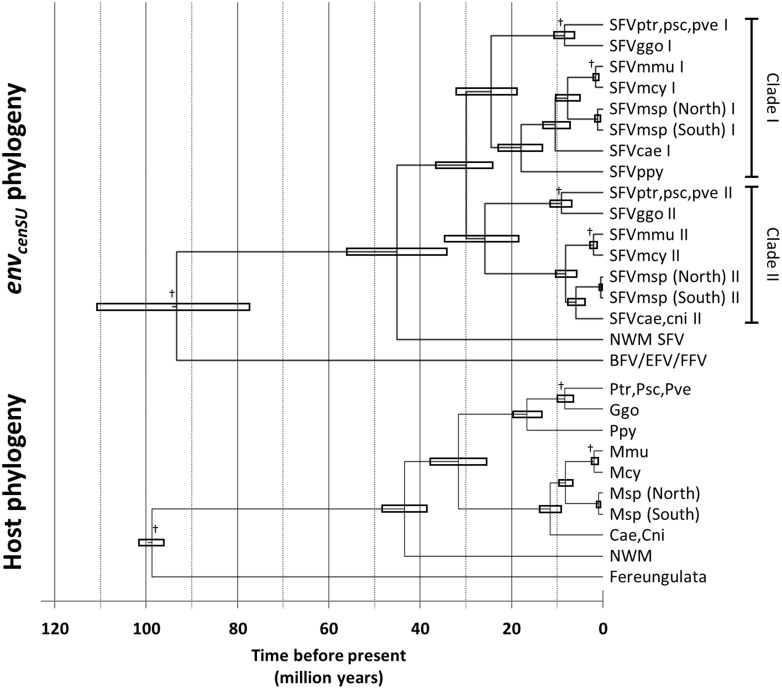
Figure 3.
Time-calibrated envcenSU (top) and host (bottom) phylogenies. The timescale of the envcenSU phylogeny was calculated under the TDRP model: , where and are time, and the corresponding number of substitutions per site, respectively, with the model parameter = 4.114 (95% HPD = 3.827–4.383), = 1.762 (95% HPD = 1.556–1.985). Sequences from fereungulata FVs were included to better estimate the TDRP model. The tree was drawn manually as the TDRP model has not yet been implemented in BEAST, and shows only nodes with important timescales we discussed in the study. The topology of the host tree and the divergence dates were estimated elsewhere (see the references in Supplementary Table S2). The node bars represent the uncertainties of the evolutionary timescale. The time is in the units of million years. The symbol ‘†’ denotes the nodes used in the TDRP model calibration. The estimated divergence dates of viruses and their hosts, as well as associated uncertainties, can be found in Supplementary Table S2. SFV, simian foamy virus; Psc, Pan troglodytes schweinfurthii chimpanzee; Pve, Pan troglodytes verus chimpanzee; Ptr, Pan troglodytes troglodytes chimpanzee; Ggo, Gorilla gorilla gorilla gorilla; Ppy, Pongo pygmaeus orangutan; Mmu, Macaca mulatta macaque; Mcy, Macaca cyclopis macaque; Msp, Mandrillus sphinx mandrill; Cae, Chlorocebus aethiops grivet; Cni, Cercopithecus nictitans greater spot-nosed monkey; NWM, New World Monkey; BFV, bovine foamy virus; EFV, equine foamy virus; FFV, feline foamy virus.