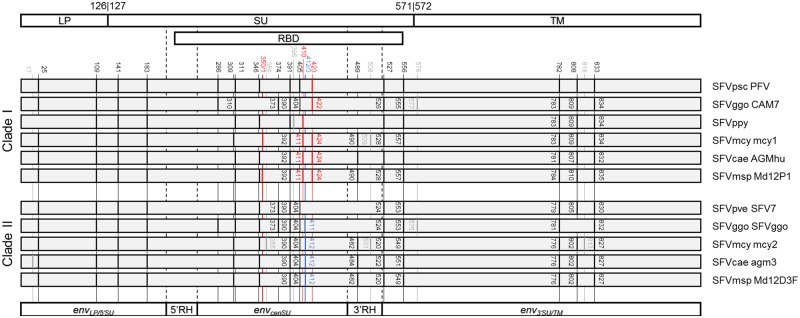
Figure 4.
N-glycosylation sites on the Env proteins. Top, schematic outline of the Env domain structure. The numbers shown indicate the beginning and the end of the domains with respective to that of the PFV (accession number: CAA69004, reported in the GenBank file Y07725) (Duda et al. 2006). Middle, N-glycosylation pattern on SFV Env proteins. N-glycosylation sites were predicted using NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) for eleven sequences. Six are those belonging to clade I, and five are those in Clade II. Sites that differ by only one amino acid are annotated together. Black lines indicate those that are shared between the two variants. Red lines indicate those that are present only in variant I sequences, and blue lines mark sites that are unique to variant II sequences. N-glycosylation sites that are unique to one host-specific viral group are marked with grey lines. The numbers are the locations of the predicted N-glycosylation sites in each protein sequence. Bottom, schematic outline of the envLP/5′SU, envcenSU, enu3′SU/TM, and RH regions of the env gene. LP, leader peptide; SU, surface domain; TM, transmembrane domain; RBD, receptor binding domain; RH, recombination hotspot; SFV, simian foamy virus; Psc, Pan troglodytes schweinfurthii chimpanzee; Pve, Pan troglodytes verus chimpanzee; Ggo, Gorilla gorilla gorilla gorilla; Ppy, Pongo pygmaeus orangutan; Mcy, Macaca cyclopis macaque; Cae, Chlorocebus aethiops grivet; Msp, Mandrillus sphinx mandrill.