Figure 1.
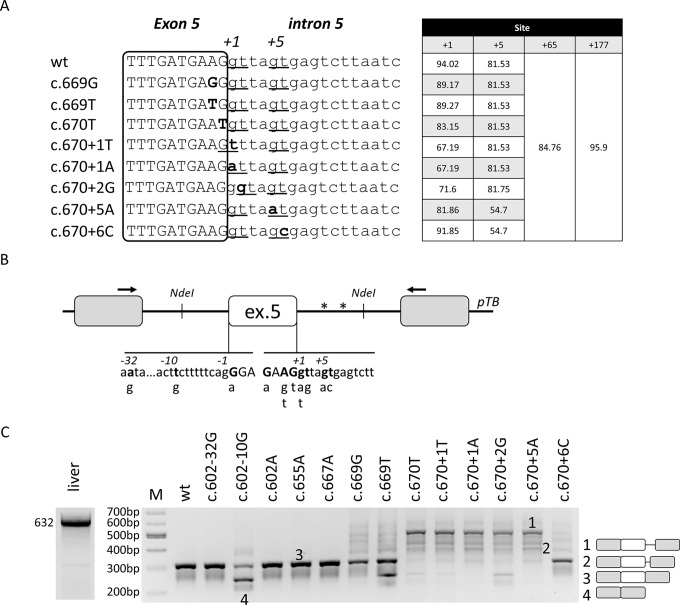
Nucleotide variants of F8 exon 5 induce aberrant splicing, ranging from exon skipping to cryptic 5’ss usage.(A) Bioinformatic analysis of 5’ss in the wild-type context or upon introduction of nucleotide changes reported into the HA mutation database at www.factorviii-db.org/ and https://databases.lovd.nl/shared/genes/F8. Their score is based on HFR matrix according to the Human Splicing Finder online software (www.umd.be/HSF/). Sequences of exon (boxed) and intron 5 are indicated respectively in upper and lower cases. Nucleotide changes are indicated in bold and the predicted 5’ss are underlined, with the relative scores reported on the right. (B) Schematic representation of the F8 exon 5 minigene cloned into the pTB vector. Exonic and intronic sequences are represented by boxes and lines, in upper and lower cases, respectively. Nucleotides reported in HA patients, together with their relative nucleotide changes, are indicated in bold and in the lower part of the figure. Asterisks represent cryptic 5’ss located at position +65 and +177 in intron 5. (C) Evaluation of F8 alternative splicing patterns in HEK293T cells transiently transfected with minigene variants. The schematic representation of the transcripts (with exons not in scale) is reported on the right. Numbers represent respectively the transcripts with +176 (1) and +64 (2) intronic nucleotides, wild-type transcripts (3), or those missing exon 5 (4). Amplified products were separated on 2% agarose gel. M, 100 bp molecular weight marker. Amplification of mRNA spanning exon 4 through exon 8 in human liver cDNA is reported on the left.