Abstract
Background
Polycomb repressive complex 2 (PRC2) is an epigenetic transcriptional repression system, whose catalytic subunit (ENHANCER OF ZESTE HOMOLOG 2, EZH2 in animals) is responsible for trimethylating histone H3 at lysine 27 (H3K27me3). In mammals, gain-of-function mutations as well as overexpression of EZH2 have been associated with several tumors, therefore making this subunit a suitable target for the development of selective inhibitors. Indeed, highly specific small-molecule inhibitors of EZH2 have been reported. In plants, mutations in some PRC2 components lead to embryonic lethality, but no trial with any inhibitor has ever been reported.
Results
We show here that the 1,5-bis (3-bromo-4-methoxyphenyl)penta-1,4-dien-3-one compound (RDS 3434), previously reported as an EZH2 inhibitor in human leukemia cells, is active on the Arabidopsis catalytic subunit of PRC2, since treatment with the drug reduces the total amount of H3K27me3 in a dose-dependent fashion. Consistently, we show that the expression level of two PRC2 targets is significantly increased following treatment with the RDS 3434 compound. Finally, we show that impairment of H3K27 trimethylation in Arabidopsis seeds and seedlings affects both seed germination and root growth.
Conclusions
Our results provide a useful tool for the plant community in investigating how PRC2 affects transcriptional control in plant development.
Keywords: Arabidopsis, PRC2, H3K27me3, EZH2 inhibitor, H3K4me3
Background
Polycomb group (PcG) proteins are a transcriptional repression system for the epigenetic control of cell, tissue and organ differentiation, contributing to correctly attain a variety of plant developmental programs. PcG proteins are grouped into two complexes: POLYCOMB REPRESSIVE COMPLEX 1 (PRC1) and 2 (PRC2). PRC2 is responsible for the trimethylation of lysine 27 of histone3 (H3K27me3), which is recognized by PRC1 to establish a silent chromatin conformation by monoubiquitination of histone H2A [1]. Furthermore, it has been demonstrated that PRC1 activity can also be required for PRC2 recruitment, and that both complexes can function independently [2–4]. Numerous studies have shown that PRC2 and, in turn, H3K27me3 play crucial roles in cell fate determination during development both in animals and plants [5, 6]. More recently, it has been shown that the two Arabidopsis homologous proteins SHORT LIFE (SHL) and EARLY BOLTING IN SHORT DAYS (EBS), following interaction with EMBRYONIC FLOWER 1 (EMF1), are able to recognize the H3K27me3 mark and trigger the repressive state of PRC2 target loci, thus mediating genome-wide transcriptional silencing [7, 8].
PRC2 consists of four subunits, an histone methyltransferase, a WD40 domain protein, a Zn-finger protein and a nucleosome-remodeling protein, first identified in Drosophila where they are respectively encoded by the genes ENHANCER OF ZESTE (E(Z)), EXTRA SEX COMBS (ESC), SUPPRESSOR OF ZESTE 12 (SU(Z)12), and NUCLEAR REMODELING FACTOR (NURF55) [9].
In Arabidopsis, there are 12 homologs of the Drosophila PRC2 subunits and, in particular, the histone methyltransferase EZ is encoded by three homologs (CURLY LEAF, MEDEA and SWINGER; CLF, MEA and SWN), which share a highly conserved SET domain, responsible of the catalytic activity [10]. Different combinations of the four subunits result in three PRC2-like complexes: the EMBRYONIC FLOWER (EMF), VERNALIZATION (VRN) and FERTILISATION INDEPENDENT SEED (FIS), which function in different developmental processes, although sharing some target genes [11, 12].
In flowering plants, the activity of PRC2 is crucial during endosperm formation as it controls the imprinting of several genes, and mutations in the imprinting machinery lead to embryonic lethality [13]. This has severely hindered studies on the function of PRC2 during seed development. An exception is represented by the genetic strategy used by Bouyer and collaborators, who were able to bypass the female gametophytic defect of the fertilization independent endosperm (fie) mutant through pollination of heterozygous fie mother plants with pollen from a cdka;1-fie double heterozygous line. This allowed to generate viable homozygous fie mutants, derived from seeds where the endosperm was of uniparental (maternal) origin [14, 15].
In mouse, overexpression of ENHANCER OF ZESTE HOMOLOG 2 (EZH2) and the consequent high level of H3K27me3 are hallmarks of several cancers, making EZH2 an ideal therapeutic target [16]. The first compound described as inhibitor of EZH2 was the 3-deazaneplanocin A (DZNep), which was shown to reduce H3K27me3 levels through depletion of EZH2 protein level, although with a fairly low specifity [17]. Subsequently, efforts in producing selective inhibitors of EZH2 by means of high-throughput screenings have been highly promising [18–21]. Among the compounds identified, the dual inhibitor of EZH2/EZH1, UNC1999, has been shown to be highly effective in vitro on both wild-type and both gain- and loss-of-function mutant EZH2. UNC1999 was shown to be able to reduce H3K27me3 levels as well as cell proliferation in a large number of cancer cells, without affecting EZH2 protein level [22, 23]. UNC1999 is representative of a family of inhibitors characterized by a 2-pyridone moiety; another class of selective inhibitors of EZH2, characterized by two benzylidene moieties, were generated and subsequently modified to produce a series of more specific compounds [24, 25].
Remarkably, a pharmacological approach has never been tested on plants, although it may represent a good alternative for the study of PRC2 function. Taking advantage of the homology of the PCR2 catalytic subunities of animals and plants, we have assessed the efficacy of 1,5-bis (3-bromo-4-methoxyphenyl)penta-1,4-dien-3-one (RDS 3434) compound, which belongs to the class of inhibitors characterized by two benzylidene moieties. RDS 3434 has been shown to be specifically active only against EZH2, and to be a selective EZH2 inhibitor in human leukemia cells where it induced heavy cell death in a dose-dependent manner, coupled with a reduction of H3K27me3 levels, without affecting EZH2 protein level [24].
The results we present here indicate the efficacy of the RDS 3434 compound as EZH2 inhibitor on Arabidopsis seeds, thus providing a new powerful tool in studying PRC2 action in plants.
Results
Treatment of seeds with the RDS 3434 inhibitor reduces H3K27me3 levels in Arabidopsis seedlings
The RDS 3434 inhibitor (Fig. 1) has been shown to be specifically active against EZH2 in human leukemia cells, where it induced heavy cell death in a dose-dependent manner [24]. To assess the efficacy of the RDS 3434 inhibitor on Arabidopsis seeds, we grew wild-type seeds on a medium supplied with increasing concentrations of RDS 3434 (30, 60, 120 μM), or with its solvent DMSO (control), for 5 days.
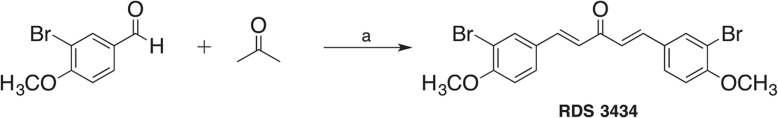
Fig. 1.
Synthesis and chemical structure of compound RDS 3434. Reagents and conditions: montmorillonite K-10, 100 W, 100 °C, 5 min, 51% yield
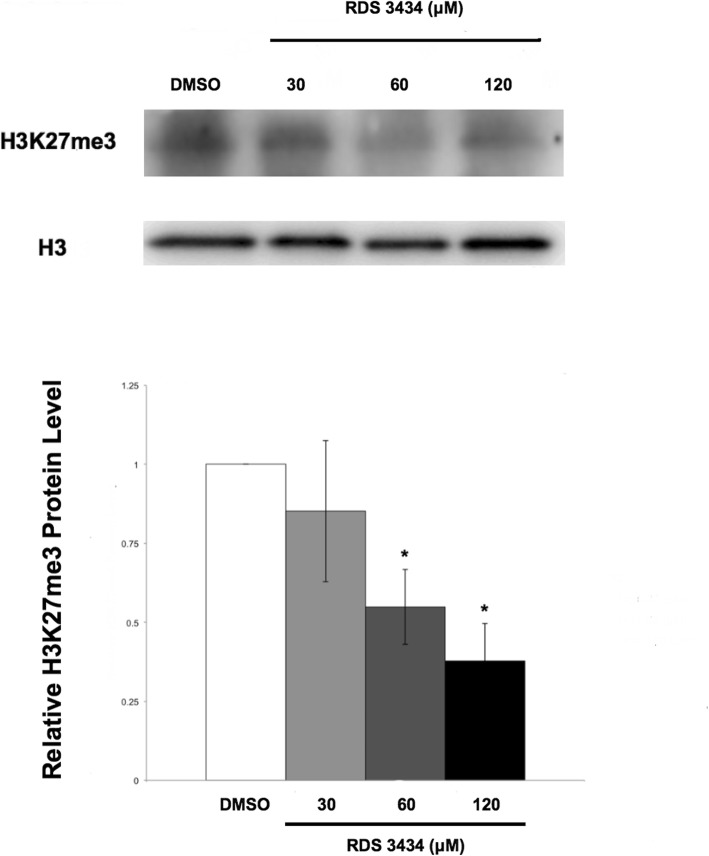
Immunoblot analysis of total proteins of RDS 3434- or DMSO-treated 5 days-old seedlings was performed with specific antibodies against H3K27me3. Measurement of the amount of proteins marked by H3K27me3 showed that the RDS 3434 inhibitor was effective in a dose-dependent manner: while with 30 μM RDS 3434 the slight decrease (16%) of H3K27me3 marked proteins compared to the control was not significant, at 60 and 120 μM they were reduced by, respectively, 45 and 62% (Fig. 2). CLF is one of the two EZH2 enzymes that play a crucial role during Arabidopsis seedling development, therefore we wondered whether the addition of the inhibitor could further affect H3K27me3 levels in a clf mutant. An immunoblot of DMSO- and RDS 3434 (120 μM)-treated clf-29 mutant seedlings compared to the DMSO- and RDS 3434-treated wild-type (Col) was performed. This analysis revealed that treatment with the inhibitor reduced by 30% the amount of proteins marked by H3K27me3 RDS 3434-treated clf seedlings (Additional file 1: Figure S1a), thus corroborating our results. In addition, the ratio of DMSO-treated clf-29/WT H3K27me3 protein level confirmed the decrease of H3K27me3 level in clf-29 compared to the wild-type (Additional file 1: Figure S1b, right).
Fig. 2.
Treatment with RDS 3434 results in a dose-dependent decrease of the total amount of H3K27me3 marked proteins. Immunoblot of 5 days-old wild-type (Ws-4) seedlings directly grown with increasing concentrations (30, 60, 120 μM) of RDS 3434 or DMSO as control. Total proteins were probed with H3K27me3 specific antibodies, and H3 was used as loading control. Western blot (top) and densitometric analysis (bottom). The protein levels are the mean of three biological replicates, presented with SD values. Significant differences were analyzed by t-test (*P ≤ 0.05, **P ≤ 0.01)
The RDS 3434 inhibitor is selectively active against the PRC2 (EZH2) complex
It is known that, in analogy with animal systems, in Arabidopsis the function of the PcG complex is counteracted by the Trithorax Group (TrxG) complex, which catalyzes the trimethylation of lysine 4 of histone 3 (H3K4me3) [26]. We thus verified whether treatment with RDS 3434 caused not only a reduction of the H3K27me3 repressive mark, but also an increase of the H3K4me3 activator mark.
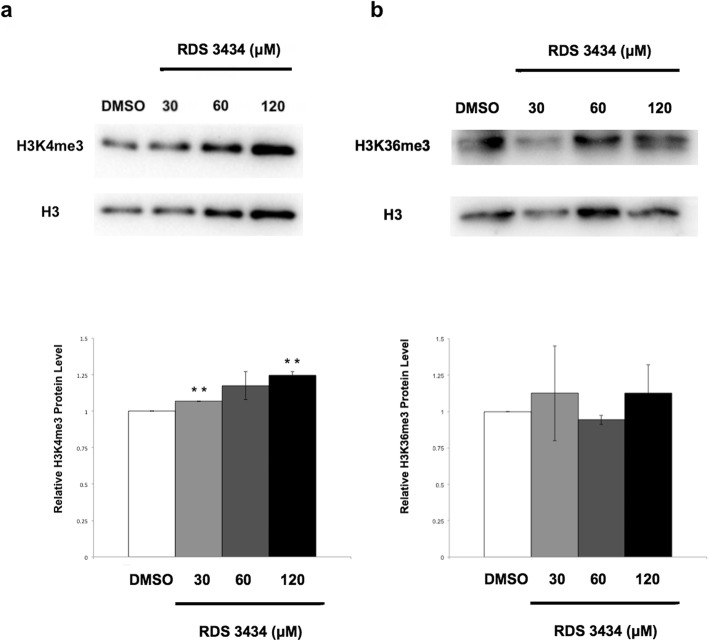
Immunoblot analysis with specific antibodies against H3K4me3 revealed that treatment with RDS 3434 produced, compared to the control, a small but significant increase in the total amount of H3K4me3 marked proteins at 30 and 120 μM inhibitor (Fig. 3a), consistent with the notion that these antagonistic marks are, in small part, mutually exclusive [27, 28].
Fig. 3.
Reduction of the H3K27me3 mark causes an increase of the antagonistic mark H3K4me3. a, b Immunoblot of 5 days-old wild-type (Ws-4) seedlings directly grown for 5 days in the presence of increasing concentrations (30, 60, 120 μM) of RDS 3434 or DMSO as control. Total proteins were probed with: H3K4me3 (a) or H3K36me3 (b) specific antibodies. H3 was used as loading control. Western blot (top) and densitometric analysis (bottom). The protein levels are the mean of two biological replicates, presented with SD values. Significant differences were analyzed by t-test (*P ≤ 0.05, **P ≤ 0.01)
To demonstrate that RDS 3434 inhibition was specific for the PRC2 (EZH2) complex over other methyltransferases, we performed an immunoblot analysis with antibodies against H3K36me3, an activating epigenetic mark catalyzed by the SET DOMAIN GROUP 8 (SDG8) [29]. This analysis showed that the H3K36me3 total protein level was not significantly affected even at the highest concentration of RDS 3434 (Fig. 3b), thus suggesting that this inhibitor functions only on the EZH2 metyltransferase.
Treatment with RDS 3434 increases the expression level of PRC2 target genes
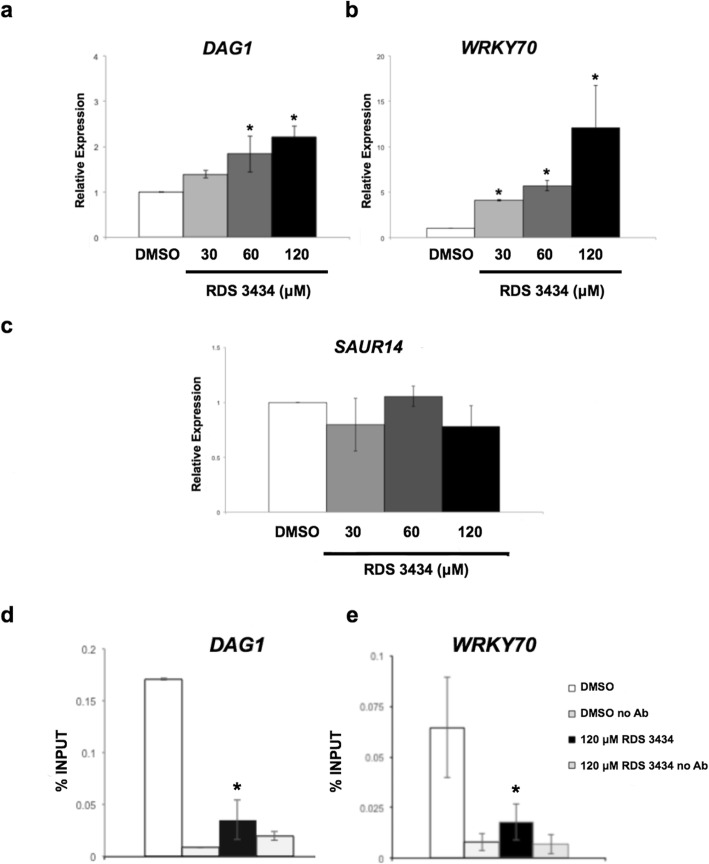
Since PRC2 is a transcriptional repression system, inhibition of EZH2 and consequent decrease of H3K27me3 levels should result in the transcriptional derepression of PRC2 target genes. Thus, we assessed whether treatment with the RDS 3434 inhibitor would actually affect the expression of two indipendent Arabidopsis PRC2 target genes: DOF AFFECTING GERMINATION 1 (DAG1), and WRKY70, respectively encoding a Dof and a WRKY transcription factor (TF). DAG1 encodes a negative regulator of seed germination [30–32], which is marked by H3K27me3 in seeds and seedlings [33] and had been shown to be upregulated in mutant plants lacking PRC2 [14].
WRKY70 encodes a TF involved in the cross-talk between salicylic acid- and jasmonic acid-dependent defense signaling, and has been reported to be a target of both PRC2 and Trithorax (Trx) [34, 35]. We performed an expression analysis (RT-qPCR) on RNA extracted from RDS 3434-treated (30, 60,120 μM) and DMSO-treated 5 days-old seedlings.
As shown in Fig. 4a and b, the expression level of both DAG1 and WRKY70 significantly increased upon treatment with RDS 3434 in a dose-dependent fashion - respectively 1.9- and 2.3-fold the level of the control for DAG1 (60 and 120 μM RDS 3434), and 4.2-, 5.8- and 12.1-fold for WRKY70 (30, 60,120 μM RDS 3434). Under the same experimental conditions, the expression level of SMALL AUXIN UP RNA 14 (SAUR14), which is not a PRC2 target gene [36], was not affected by treatment with RDS 3434, thus confirming the efficacy of this inhibitor only for PRC2 (Fig. 4c). We then assessed whether treatment with the inhibitor would actually result in loss of the H3K27me3 repressive mark in the PRC2 target loci DAG1 and WRKY70. To this end, we performed chromatin immunoprecipitation (ChIP) assays with H3K27me3-specific antibodies, or without antibodies as negative control, on samples derived from RDS 3434 (120 μM)-treated and DMSO-treated 5 days-old seedlings. We measured the enrichment of H3K27me3 by amplification, through quantitative (qPCR), of one region in the body of both DAG1 and WRKY70 genes, because the H3K27me3 epigenetic mark is usually restricted to the transcribed regions of target genes [37, 38]. Interestingly, in samples derived from RDS 3434 (120 μM)-treated seedlings, the level of H3K27me3 in the DAG1 and WRKY70 genes was significantly decreased (Fig. 4d, e), consistently with their increased expression levels.
Fig. 4.
Inhibition of EZH2 results in an increased expression of two PRC2 target genes. a, c Relative expression level of the PRC2 target genes, DAG1 (a) and WRKY70 (b), and of the non-target gene SAUR14 (c), in wild-type (Ws-4) seedlings directly grown for 5 days in the presence of increasing concentrations (30, 60, 120 μM) of RDS 3434 or DMSO as control. Relative expression levels were normalized with that of the GAPDHa (At3g26650) gene, and are presented by the ratio of the corresponding mRNA level in the control, which was set to 1. d, e Chromatin from samples derived from 5-days old seedlings grown in the presence of 120 μM RDS 3434 or DMSO as control, immunoprecipitated with anti-H3K27me3 antibodies, or without antibodies as a negative control. The amount of DNA for DAG1 (d) or WRKY70 (e) was measured by qPCR. The values of fold enrichment were normalized to input. All the primers used are listed in Table 1. The results were obtained from two independent replicates with SD values. Significant differences were analyzed by t-test (*P ≤ 0.05, **P ≤ 0.01)
Reduction of the H3K27me3 mark affects seed germination and root development
The transcriptional control mediated by PRC2 is crucial during seed germination, as it silences seed specific genes thus allowing proper seedling growth and development [14, 26]. Therefore, we assessed whether treatment with the inhibitor RDS 3434 would affect germination of seeds. As shown in Fig. 5a, treatment with the inhibitor caused a significant reduction of the germination rate at 24 h after imbibition (HAI) - 26.5 and 34.3%, at 60 and 120 μM RDS 3434, respectively.
Fig. 5.
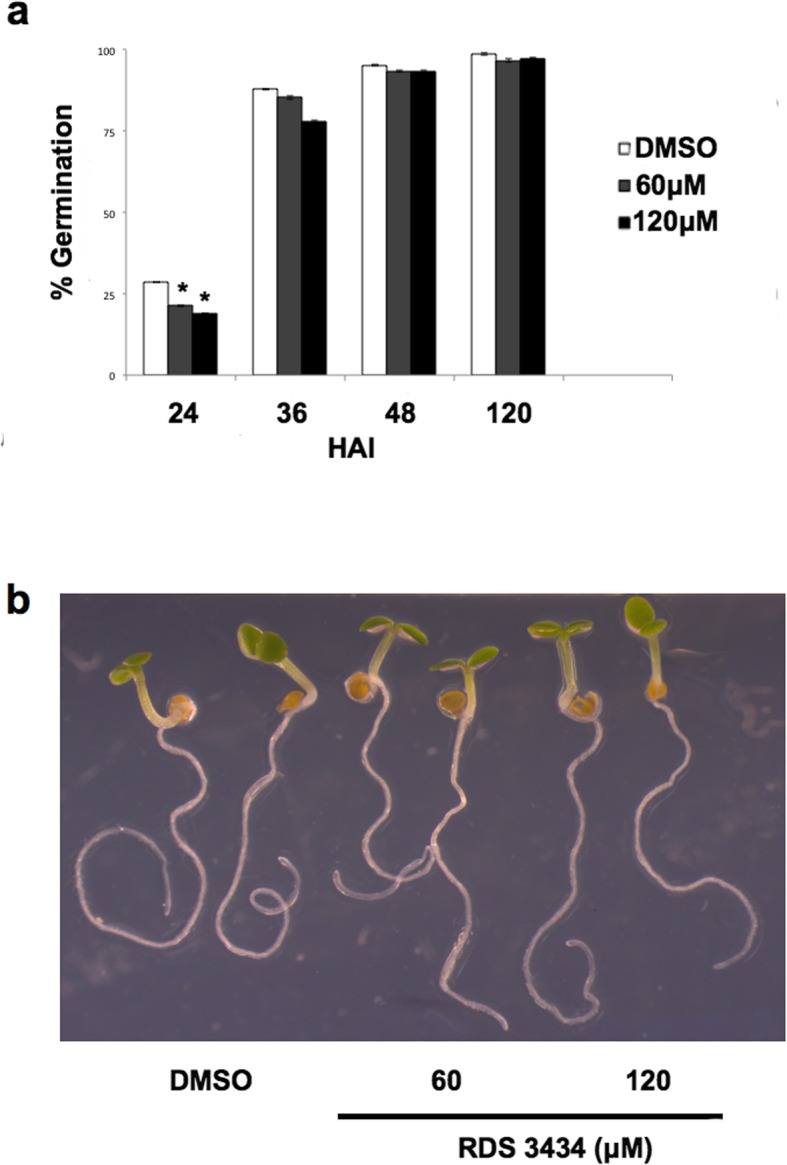
Inhibition of EZH2 results in delayed seed germination. a Seed germination assays of wild-type (Ws-4) seeds imbibed in the presence of RDS 3434 (60, 120 μM) or DMSO as control. Germination rate was scored at 24, 36, 48 and 120 HAI (Hours After Imbibition). Data represent the mean of two independent biological replicates each performed in duplicate (25 seeds per replica). Significant differences were analyzed by t-test (*P ≤ 0.05, **P ≤ 0.01). b 5 days-old wild-type (Ws-4) seedlings directly grown for 5 days in the presence of increasing concentrations (60, 120) of RDS 3434 or DMSO as control
On the other hand, seedling growth of treated and untreated samples was very similar (Fig. 5b), ruling out the possibility that the observed reduction of H3K27me3 upon treatment with RDS 3434, may pleiotropically affect seedling growth and development. Similarly, 5 days-old seedlings of clf or swn single mutants do not show severe developmental defects, since only clf adult plants are characterized by dwarfism and early flowering, whereas swn mutants display very weak phenotypes [39, 40]. Since treatment with RDS 3434 further reduces H3K27me3 level in clf-29 seedlings, we assessed whether seed germination of mutant seeds would be affected by treatment with RDS 3434 (120 μM). Interestingly, clf-29 mutant seeds treated with the inhibitor showed a 50% reduction of the germination rate at 24 HAI (Additional file 2: Figure S2a). However, treatment with the inhibitor did not result in more severe phenotypes during seedling development (Additional file 2: Figure S2b).
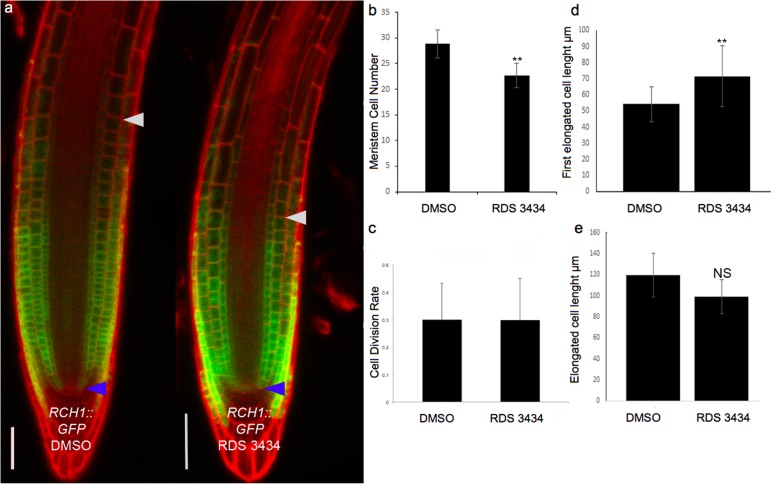
It has been proposed that PRC2 controls primary root growth, since lack of the EZ catalytic subunits CLF and SWN results in short meristem and decreased primary root length [41]. Therefore, we assessed whether treatment with the RDS 3434 inhibitor results in root developmental defects. Although treatment with 60 and 120 μM RDS 3434 inhibitor did not affect root growth (data not shown), a higher dose (240 μM) resulted in a reduced number of root meristematic cells. Consistently, the expression domain of ROOT CLAVATA HOMOLOG1 (RCH1), a gene specifically marking the root meristematic zone [42], is reduced in RDS 3434-treated plants compared to the control, as visualized by a RCH1- GFP transcriptional fusion (Fig. 6a, b).
Fig. 6.
Inhibition of EZH2 affects root development. a Confocal microscopy images of RCH1::GFP roots from 5 days-old seedlings grown in the presence of RDS 3434 (240 μM) or DMSO as control. Blue and white arrowheads indicate the Quiescent Center (QC) and the cortex Transition Boundary (TB), respectively. b Root meristem cell number. c Quantification of GUS spots per meristem in treated and untreated of CYCLINB1;1pro:CDB-GUS roots. d, e Length of the first elongated cell (d), and of the differentiated cell (e) (n = 30). Data represent the mean of two independent biological replicates, presented with SD values. Significant differences were analyzed by t-test (*P ≤ 0.05, **P ≤ 0.01)
A decrease in meristem size can be caused by a reduced division rate or by a more rapid elongation/differentiation (i.e. exit from the meristem) of meristematic cells. To distinguish between these two possibilities, we first visualized root meristem cells in the G2–M phase in RDS 3434-treated and untreated plants harboring the D-Box CYCB::GUS construct, a marker of the G2-M transition [43]: no difference in the cell division potential could be detected (Fig. 6c). To detect possible variation in cell elongation/differentiation, we measured the length of the first elongated and of the fully differentiated cells in both treated and untreated plants. Whereas the length of fully elongated cells was unvaried, the first elongated cells were longer in RDS 3434-treated plants (Fig. 6d, e), indicating that RDS 3434 affects root meristem activities controlling the elongation/differentiation potential. On the other hand, the overall appearance of 240 μM-treated seedling was similar to that of DMSO-treated seedlings (Additional file 3: Figure S3).
Discussion
In this work, we describe a simple methodology for inhibiting in plants the methyltransferase activity of EZH2, the catalytic subunit of the PRC2 complex, and report the effects of this inhibition on Arabidopsis seed germination and root growth.
PRC2 plays a crucial role in the embryo-to-seedling [14, 44, 45] and vegetative-to-reproductive developmental phase transitions [46, 47]. PRC2 is also essential for endosperm formation, since it controls the parent-of-origin specific expression of a number of genes: lack of the maternal PRC2 function results in derepression of target genes, causing endosperm overproliferation and eventually seed abortion [13, 48]. This has severely hampered studies on the function of PRC2 during seed development.
In mammals, studies on PRC2 function have benefited of the development and use of inhibitors of the catalytic subunit EZH2, among which RDS 3434 whose effectiveness has been proven on the oncogenic human monocyte cell line U937 [24]. Since the catalytic subunit EZH2 is highly conserved between mammals and plants, we tested the effectiveness of RDS 3434 in Arabidopsis and found that indeed it inhibits PRC2-mediated H3K27me3 methylation also in this organism.
Our study does not indicate whether inhibition of methylation is specific for a particular H3K27me3 methyltransferase of the Arabidopsis E(z) group, namely CLF, SWN or MEA, although our data concern the seed-to-seedling transition where CLF and/or SWN are active [11]. Previous genome-wide analyses comparing the global H3K27me3 profile in clf-28 or clf-29 mutant seedlings, which revealed a decreased level of H3K27me3 in the mutant lines compared to the wild-type [49–51].
Consistently, our immunoblot analysis showed the decrease of H3K27me3 level in clf-29 compared to the wild-type, and revealed that treatment with RDS 3434 further reduces the amount of proteins marked by H3K27me3 in the clf-29 mutant background.
It will be interesting to assess the efficacy of RDS 3434 during gametophyte and endosperm development, where the EZH2 subunit involved is MEA [52].
It has been previously demonstrated that lack of both the catalytic subunits CLF and SWN result in delayed germination: clf-28 swn-4 double mutant seeds germinate within 4 DAI (Days After Imbibition), while wild-type seeds germinate within 2 DAI [14]. Similarly, seeds of the double mutant atmib1a atmib1b, which lacks the E3 ubiquitin ligase subunit BMI of the PRC1 complex [53, 54], reach full germination at 6 DAI whereas the corresponding wild-type seeds germinated at 3 DAI [26]. Consistently, treatment with RDS 3434 affects seed germination in a dose-dependent fashion, as wild-type seeds treated with RDS 3434 showed a significant reduction of the germination rate within the first 36 h compared to untreated seeds; this phenotype is even more pronounced in the clf-29 mutant background, thus corroborating the effectiveness of RDS 3434 as an inhibitor. It should be pointed up that the clf-29 single mutant has been previously characterised for the early flowering phenotype, as well as for the curly leaves and dwarf adult plants [55, 56]. Also the null clf-50 allele, regardless of the different genetic background (Ws), displayed similar phenotypes, which were enhanced by the swn-1 weak allele [46]. In addition, it has been shown that clf-29 display an increase in the number of root meristematic cells [41, 57]), conversely to the swn-7 allele which has a shorter root with no difference in meristem size [41].
The clf-28 swn-7 double mutant lacking both EZH2 subunits [41], and the fertilization independent endosperm 2 (fie) mutant, which lacks the Arabidopsis homolog of the DROSOPHILA EXTRA SEX COMBS (ESC) PRC2 subunit [14], have shorter roots and smaller meristems with fewer meristematic cells than wild-type. Consistently, seedlings treated with RDS 3434 displayed a decrease of both root meristem size and meristematic cell number, due to an effect of the inhibitor on the elongation/differentiation potential of meristematic cells.
In Drosophila and in animal stem cells the function of the PRC2 complex is counteracted by the Trithorax Group (TrxG) proteins, which catalyze the trimethylation of lysine 4 of histone 3 (H3K4me3) that acts as a transcriptional activator epigenetic mark [58–61]. In Drosophila and mammals the silencing effect of H3K27me3 is counteracted by the inductive action of H3K4me3 [60, 62]. In Arabidopsis, genome-wide analysis of H3K4me3 and H3K27me3 revealed that only a number of genes are targets of both these antagonistic chromatin marks [37, 38]. Among these genes, key regulators of flower development in the vegetative-to-reproductive transition have been shown to be transcriptionally regulated by H3K4me3 and H3K27me3 [63]. As for the seed-to-seedling transition, a switch from an activated to a repressed state associated to H3K4me3 and H3K27me3, respectively, has been reported for a number of seed developmental genes during germination and early seedling development [26, 33]. However, the antagonism between these two epigenetic marks in Arabidopis is still controversial [64] and it has been recently proposed that Trx proteins cooperate with the PRC2 proteins to repress seed-specific genes during germination and seedling development [65]. In addition, comparative analysis of wild-type and fie mutant seedlings lacking a functional PRC2 revealed a genome-wide absence of the H3K27me3 mark in the fie mutant; of the H3K27me3 mark-free PRC2 target genes in fie seedlings, only a limited number are transcriptionally induced and associated with the H3K4me3 activating mark [14].
In agreement with these results, we show that removal of the repressive mark H3K27me3 involves the establishment of the activating mark H3K4me3 only to a certain degree, since RDS 3434-treated seedlings showed only a slight, although significant, increase in the level of H3K4me3-marked proteins.
Besides being effective in Arabidopsis, we also show that the RDS 3434 inhibitor functions only on the H3K27me3 epigenetic mark; indeed, this compound does not inhibit the methyltransferases of the SET DOMAIN GROUP 8 (SDG8), which catalyzes trimethylation of H3K36 [66], as the entire bulk of H3K36me3 was not significantly different in treated samples compared to the untreated control. In addition, expression of the non-PRC2 target gene SAUR14 (see below) is unchanged in treated samples compared to the control, thus corroborating the selectivity of this inhibitor.
Conclusions
Our results indicate that RDS 3434, a compound previously characterised as inhibitor of the human EZH2 subunit, is an effective and selective inhibitor of the PRC2 complex also in plants.
This study can be of significant interest for the community investigating Polycomb activity in plants, as no chemical inhibition specific for H3K27me3 has been reported so far; RDS 3434 could represent a powerful tool to further investigate the effects of the transcriptional control mediated by PRC2 in plants.
Methods
General
Reagents and solvents employed were obtained from Aldrich. Melting point (uncorrected) of RDS 3434 was determined in open Pyrex capillary tubes using a Buchi 510 melting point apparatus. NMR spectrum of the synthesized compound was run on a Bruker Avance system, operating at 400 MHz; the solvent used for NMR analysis was DMSO-d6. Microwave reactions were conducted using a CEM Discover system unit consisting of a continuous focused microwave-power delivery system. The temperature was monitored using a calibrated infrared temperature control mounted under the reaction vessel. The contents of the vessel are stirred by means of a rotating magnetic plate located below the floor of the microwave cavity.
Synthesis of compound RDS 3434
Derivative RDS 3434 was synthesized by microwave-assisted condensation of propanone with 3-bromo-4-methoxy-benzaldehyde (Fig. 1) following a previously reported synthetic approach [67]. 3-Bromo-4-methoxy-benzaldehyde (1.5 mmol) was dissolved in MeOH and treated with montmorillonite K-10 (600 mg). Evaporation of the solvent gave a dispersion that was suspended in propanone (0.75 mmol) and irradiated at 100 W for 5 min at 100 °C. After cooling, the mixture was diluted with MeOH, filtered and evaporated to dryness. The crude was purified by column chromatography (SiO2, chloroform/n-hexane 9:1) and recrystallized from MeOH to give the pure product as a yellow solid (51% yield). mp: 154–156 °C; IR ν 1554 (C=O ketone) cm− 1; 1H NMR (DMSO d6, δ): 3.90 (s, 6H), 7.12–7.25 (m, 4H), 7.55–7.75 (m, 4H), 8.05 (s, 2H).
Plant material and growth conditions
The Arabidopsis thaliana wild-type line used in this work is Ws-4, unless specified, and was grown in a growth chamber at 24/21 °C with 16/ 8-h day/night cycles and light intensity of 300 μmol/m− 2 s− 1, as previously described [30]. Seeds were surface sterilized and plated on MS agar (halfstrength MS, 0.8% agar, pH 5.7), unless specified, and stratified at 4 °C for three days in the dark. The clf-29 mutant line is in Col-0 (SALK_N521003), and was kindly provided by Dr. Miguel de Lucas. The marker lines RCH1:: GFP and CYCLINB1;1pro:CDB-GUS are in Col-0 ecotype and were previously described [68, 69]. The corresponding wild type line was used for the root analysis. The wild-type lines (Ws-4 and Col-0) have been obtained from the European Arabidopsis Stock Centre (arabidopsis.info). As for the treatment with RDS 3434, wild-type seeds were sown on medium supplied with increasing concentrations of RDS 3434 (30, 60, 120 μM), or with an equal volume of its solvent DMSO (Dimethyl sulfoxide), as control.
Protein extraction and Immunoblot analysis
Five days-old seedlings, grown in the presence of increasing concentrations of RDS 3434 (30, 60, 120 μM), or with an equal volume of its solvent DMSO as controls, were grinded with liquid nitrogen and dissolved in Chromatin Buffer Extraction (Sucrose 0,4 M; Tris Hcl pH 8 10 mM; β-mercaptoethanol 5 mM; PMSF 0,1 mM; Protease inhibitor cocktail 1X, Sigma-Aldrich P9599). The nuclei were pelleted at 4000 rpm for 20 min, at 4 °C, and dissolved in 800 μl ddH2O. The proteins were precipitated with 150 μl of NaOH/ β-mercaptoethanol (138,7 μl 2 N NaOH and 11,25 μl β-mercaptoethanol) and then with 55% TCA solution, for 15 min in ice. Following centrifugation for 20 min at 14000 rpm, 4 °C, the pellet was dissolved in HU Buffer (Urea 8 M; SDS 5%; TrisHCl pH 6,8200 mM; EDTA 0,1 mM; DTT 100 mM; bromophenol blu) and boiled for 10 min at 65 °C. Proteins were separated on a 12% SDS-polyacrylamide gel (Bio-Rad) and blotted on a PVDF Immobilon-P Transfer membrane (Millipore). Detection of proteins was performed with specific antibodies against H3K27me3 (Millipore #07–449), H3K4me3 (Abcam- ab8580) or H3K36me3 (Abcam-ab9050), and against histone H3 (Biorbyt orb10805) as a loading control. The anti-rabbit IgG conjugated to peroxidase was used as a secondary antibody and the signal was detected with ECL system. The values are the average of two biological replicates (except the immunoblot for H3K27me3 that was performed with three biological replicates), presented with SD values. Significant fold enrichments were analyzed by t-test (*P ≤ 0,05).
Expression analysis
Five days-old seedlings, grown in the presence of increasing concentrations of RDS 3434 (30, 60, 120 μM) or with an equal volume of its solvent DMSO as control, were frozen and grinded with liquid nitrogen. Total RNA was extracted and purified according to Lorrai et al. [70]. RT-qPCR assays were performed according to [31]. Relative expression levels were normalized with the GAPDH (At3g26650) reference gene, which is not marked by H3K27me3, and are presented by the ratio of the corresponding mRNA level of the DMSO-treated sample, which was set to 1. The primers used are listed in Table 1. Two independent biological replicates were performed, and one representative experiment is reported with SD values. Significant differences were analyzed by t-test (*P ≤ 0.05; **P ≤ 0.01).
Table 1.
Primers used in this study
| Gene name | Forward | Reverse |
|---|---|---|
| DAG1 | TTGTCGAAGGTATTGGACCGA | CCGACTGGGACGTTACGAAG |
| DAG1 ChIP | CGCAACAACAACCAACATTC | GCCGTGTTGTTGGTATTTCC |
| WRKY70 | CAGGCCAGTTACGTCAATGGGAAAA | GAAATCGCCGCCACCTCCA |
| SAUR14 | GTAGTTCCGGTTTCGTACTTGGACC | CTGCAAGGGATTGTGAGGCCA |
| GADPH | GCTGAGGAAGTCAACGCTGC | CGGACACTAGTGGCTCATCG |
Chromatin Immunoprecipitation (ChIP) assay
ChIP assay was performed according to Lorrai et al. [70]. To analyse the epigenetic profile of the DAG1 and WRKY70 loci, chromatin from 5 days-old seedlings, grown in the presence of RDS 3434 120 μM or with an equal volume of its solvent DMSO as control, was immunoprecipitated overnight using antibodies against H3K27me3 (Millipore #07–449), or without antibodies as negative control. After reverse cross-linking, the enriched DNA levels were quantified by qPCR using specific primer sets listed in Table 1. The fold enrichment of a specific region was normalized for the Input fraction, to minimize the background differences among the sample. Two replicates were performed, and one representative experiment is reported with SD values. Significant fold enrichments were analyzed by t-test (*P ≤ 0,05).
Seed germination assay
All seeds used for germination tests were harvested from mature plants grown at the same time, in the same conditions, and stored for 4–5 weeks in the dark under dry conditions at room temperature. For seed germination assays, triplicate sets of 25 seeds were surface sterilized and plated on agar (0.8%) with increasing concentrations of RDS 3434 (60, 120 μM) or with an equal volume of its solvent DMSO as control. Germination rate was scored at 24, 36, 48 and 120 h after imbibition (HAI). Germination rate was calculated as the ratio of seeds with the radicle protruding from the seed coat on the total number of seeds sown (n = 25). The values are the mean of two biological replicates presented with SD values. Significant differences were analyzed by t-test (*P ≤ 0.05; **P ≤ 0.01).
Root meristem size analysis
Root analyses were performed on roots from five days-old seedlings, grown on MS agar supplemented with 0.5% sucrose for 3 days, then transferred to the same medium in the presence of RDS 3434 (240 μM) or with an equal volume of its solvent DMSO as control, for 2 days. For light DIC microscopy, samples were mounted on a media containing chloral hydrate (SigmaAldrich): 3 parts glycerol: 1 parts water. Images were acquired utilizing Nomarski optics under a Zeiss Axio Imager.A2 microscope with a dry 40X objective. For all the analyses, at least 30 samples were analyzed and statistically treated. Root meristem size was measured based on the number of cortex cells in a file extending from the quiescent center to the first elongated cortex cell excluded as previously described [69]. Images were obtained using a confocal laser scanning microscope (Zeiss LSM 780). The length of both the first elongated cell and differentiated cell was measured using image J. Significant differences were analyzed by t-test (*P ≤ 0.05; **P ≤ 0.01).
Statistical analysis
Each experiment was performed in duplicate and repeated with two or three independent biological replicates. Results are expressed as mean (except for the expression analysis) ± standard deviation (SD). Two-tailed Student’s t-test was used to evaluate statistical significance, respect to the DMSO control (*P ≤ 0.05; **P ≤ 0.01).
Supplementary information
Additional file 1. Figure S1. Treatment with RDS 3434 reduces H3K27me3 protein level in the curly leaf-29 mutant.a, b Immunoblot of 5 days-old clf-29 seedlings directly grown with RDS 3434 (120 μM) or DMSO as control (a), and of DMSO- or RDS 3434-treated wild-type (Col) and clf-29 seedlings (b). Total proteins were probed with H3K27me3 specific antibodies, and H3 was used as loading control. Western blot (top) and densitometric analysis (bottom). In (b) is shown the relative H3K27me3 protein level (bottom left), and the ratio of DMSO- and 120 μM RDS 3434-treated clf-29/WT (bottom right). Results were obtained from two independent replicates with SD values. Significant differences were analyzed by t-test (*P ≤ 0.05).
Additional file 2. Figure S2. Treatment with RDS3434 of clf-29 mutant seeds affects seed germination. a Seed germination assays of clf-29 mutant seeds imbibed in the presence of RDS 3434 (120 μM) or DMSO as control. Germination rate was scored at 24, 36, 48 and 120 HAI (Hours After Imbibition). Data represent the mean of two independent biological replicates each performed in duplicate (25 seeds per replica). Significant differences were analyzed by t-test (*P ≤ 0.05, **P ≤ 0.01). b 5 days-old clf-29 mutant seedlings directly grown for 5 days in the presence of RDS 3434 (120 μM) or DMSO as control.
Additional file 3. Figure S3. Wild-type seedlings treated with 240 μM RDS 3434. 5 days-old wild-type (Ws-4) seedlings directly grown in the presence of of RDS 3434 (240 μM) or DMSO as control.
Acknowledgements
We thank Prof. Rodolfo Negri for constructive suggestions on this work, and Andrea Lepri for his helpful technical assistance. We also thank Miguel de Lucas and Marie-Terese Hauser for providing seeds of curlyleaf29 (clf-29) and CYCLINB1;1pro:CDB-GUS lines, respectively.
Authors’ informations
AB Present Address: Center for Integrative Genomics, Faculty of Biology and Medicine, University of Lausanne, CH-1015 Lausanne, Switzerland.
Abbreviations
- CLF
CURLY LEAF
- CYCB::GUS
CYCLINB1;1pro:CDB-GUS
- DAG1
DOF AFFECTING GERMINATION1
- DMSO
Dimethyl sulfoxide
- DZNep
3-deazaneplanocin
- E(Z)
ENHANCER OF ZESTE
- EBS
EARLY BOLTING IN SHORT DAYS
- EMF
EMBRYONIC FLOWER
- EMF1
EMBRYONIC FLOWER 1
- ESC
EXTRA SEX COMBS
- EZH2
ENHANCER OF ZESTE HOMOLOG 2
- fie
Fertilization independent endosperm
- FIS
FERTILISATION INDEPENDENT SEED
- H3K27me3
Histone H3 at lysine 27 trimethylated
- H3K4me3
Histone H3 at lysine 4 trimethylated
- HAI
Hours after imbibition
- MEA
MEDEA
- NMR
Nuclear Magnetic Resonance
- NURF55
Nuclear remodeling factor
- PcG
Polycomb group
- PRC1, PRC2
Polycomb Repressive Complex 1, 2
- RCH1
ROOT CLAVATA HOMOLOG1
- RDS 3434
1,5-bis(3-bromo-4-methoxyphenyl)penta-1,4-dien-3-one
- RT-qPCR
Reverse transcriptase- quantitative polymerase chain reaction
- SAUR14
SMALL AUXIN UP RNA 14
- SDG8
SET DOMAIN GROUP 8
- SHL
SHORT LIFE
- SU(Z)12
SUPPRESSOR OF ZESTE 12
- SWN
SWINGER
- TrxG
Trithorax Group
- VRN
VERNALIZATION
Authors’ contributions
VR, AB and PV conceived and designed the research. PV directed the project. VR, CL and RL performed the experiments. VNM, FS, VT, RDS and RC designed and synthesized the inhibitor. RDI and SS performed root phenotypic analysis and analysed the data. PV and PC wrote the manuscript. VR, AB, SS revised the manuscript. All Authors read and approved the final manuscript.
Funding
This research was supported by research grants from Ministero dell’Istruzione, Università e Ricerca, Progetti di Ricerca di Interesse Nazionale, and from Sapienza Università di Roma to P.C., and from Sapienza Università di Roma and Istituto Pasteur-Fondazione Cenci Bolognetti to P.V. The funding bodies provided the financial support to the research project, but were not involved in the design of the study, in the collection, analysis and interpretation of data, and in writing the manuscript.
Availability of data and materials
The data sets supporting the results of this article are included within the article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Veronica Ruta, Email: veronica.ruta@uniroma1.it.
Chiara Longo, Email: longo.493622@studenti.uniroma1.it.
Alessandra Boccaccini, Email: boccaccini_alessandra@hotmail.com.
Valentina Noemi Madia, Email: valentinanoemi.madia@uniroma1.it.
Francesco Saccoliti, Email: francesco.saccoliti@uniroma1.it.
Valeria Tudino, Email: valeria.tudino@uniroma1.it.
Roberto Di Santo, Email: roberto.disanto@uniroma1.it.
Riccardo Lorrai, Email: riccardo.lorrai@uniroma1.it.
Raffaele Dello Ioio, Email: raffaele.delloioio@uniroma1.it.
Sabrina Sabatini, Email: sabrina.sabatini@uniroma1.it.
Roberta Costi, Email: roberta.costi@uniroma1.it.
Paolo Costantino, Email: paolo.costantino@uniroma1.it.
Paola Vittorioso, Email: paola.vittorioso@uniroma1.it.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-019-2057-7.
References
- 1.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8(1):9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 2.Merini W, Calonje M. PRC1 is taking the lead in PcG repression. Plant J. 2015;83(1):110–120. doi: 10.1111/tpj.12818. [DOI] [PubMed] [Google Scholar]
- 3.Forderer A, Zhou Y, Turck F. The age of multiplexity: recruitment and interactions of Polycomb complexes in plants. Curr Opin Plant Biol. 2016;29:169–178. doi: 10.1016/j.pbi.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16(11):643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20(10):1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 6.Bemer M, Grossniklaus U. Dynamic regulation of Polycomb group activity during plant development. Curr Opin Plant Biol. 2012;15(5):523–529. doi: 10.1016/j.pbi.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Fu X, Wang Y, Liu R, He Y. Polycomb-mediated gene silencing by the BAH-EMF1 complex in plants. Nat Genet. 2018;50(9):1254–1261. doi: 10.1038/s41588-018-0190-0. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Qian S, Scheid RN, Lu L, Chen X, Liu R, Du X, Lv X, Boersma MD, Scalf M, et al. EBS is a bivalent histone reader that regulates floral phase transition in Arabidopsis. Nat Genet. 2018;50(9):1247–1253. doi: 10.1038/s41588-018-0187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136(21):3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 10.Muller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev. 2009;19(2):150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 2009;25(9):414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Xiao J, Wagner D. Polycomb repression in the regulation of growth and development in Arabidopsis. Curr Opin Plant Biol. 2015;23:15–24. doi: 10.1016/j.pbi.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Kohler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 2003;22(18):4804–4814. doi: 10.1093/emboj/cdg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, Goodrich J, Renou JP, Grini PE, Colot V, et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 2011;7(3):e1002014. doi: 10.1371/journal.pgen.1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowack MK, Shirzadi R, Dissmeyer N, Dolf A, Endl E, Grini PE, Schnittger A. Bypassing genomic imprinting allows seed development. Nature. 2007;447(7142):312–315. doi: 10.1038/nature05770. [DOI] [PubMed] [Google Scholar]
- 16.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1–2):21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21(9):1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu B, Konze KD, Jin J, Wang GG. Targeting EZH2 and PRC2 dependence as novel anticancer therapy. Exp Hematol. 2015;43(8):698–712. doi: 10.1016/j.exphem.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, Romeril SP, Burgess JL, Grant SW, Brackley JA, et al. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med Chem Lett. 2012;3(12):1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8(11):890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 21.Fioravanti R, Stazi G, Zwergel C, Valente S, Mai A. Six years (2012-2018) of researches on catalytic EZH2 inhibitors: the boom of the 2-Pyridone compounds. Chem Rec. 2018;18(12):1818–1832. doi: 10.1002/tcr.201800091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, Allison DF, Cai L, Rockowitz S, Liu S, et al. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2015;125(2):346–57. [DOI] [PMC free article] [PubMed]
- 23.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, Liu F, Gao C, Huang XP, Kuznetsova E, et al. An orally bioavailable chemical probe of the lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8(6):1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valente S, Lepore I, Dell'Aversana C, Tardugno M, Castellano S, Sbardella G, Tomassi S, Di Maro S, Novellino E, Di Santo R, et al. Identification of PR-SET7 and EZH2 selective inhibitors inducing cell death in human leukemia U937 cells. Biochimie. 2012;94(11):2308–2313. doi: 10.1016/j.biochi.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Ciarapica R, Carcarino E, Adesso L, De Salvo M, Bracaglia G, Leoncini PP, Dall'agnese A, Verginelli F, Milano GM, Boldrini R, et al. Pharmacological inhibition of EZH2 as a promising differentiation therapy in embryonal RMS. BMC Cancer. 2014;14:139. doi: 10.1186/1471-2407-14-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molitor AM, Bu Z, Yu Y, Shen WH. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 2014;10(1):e1004091. doi: 10.1371/journal.pgen.1004091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1(3):299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17(3):425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30(10):1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 2000;14(1):28–33. [PMC free article] [PubMed] [Google Scholar]
- 31.Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 2010;61(2):312–323. doi: 10.1111/j.1365-313X.2009.04055.x. [DOI] [PubMed] [Google Scholar]
- 32.Boccaccini A, Santopolo S, Capauto D, Lorrai R, Minutello E, Serino G, Costantino P, Vittorioso P. The DOF protein DAG1 and the DELLA protein GAI cooperate in negatively regulating the AtGA3ox1 gene. Mol Plant. 2014;7(9):1486–1489. doi: 10.1093/mp/ssu046. [DOI] [PubMed] [Google Scholar]
- 33.Boccaccini A, Lorrai R, Ruta V, Frey A, Mercey-Boutet S, Marion-Poll A, Tarkowska D, Strnad M, Costantino P, Vittorioso P. The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels. BMC Plant Biol. 2016;16(1):198. doi: 10.1186/s12870-016-0890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16(2):319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarez-Venegas R, Abdallat AA, Guo M, Alfano JR, Avramova Z. Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics. 2007;2(2):106–113. doi: 10.4161/epi.2.2.4404. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Tian T, Zhang K, You Q, Yan H, Zhao N, Yi X, Xu W, Su Z. PCSD: a plant chromatin state database. Nucleic Acids Res. 2018;46(D1):D1157–d1167. doi: 10.1093/nar/gkx919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10(6):R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5(5):e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu M, Hu T, Smith MR, Poethig RS. Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell. 2016;28(1):28–41. doi: 10.1105/tpc.15.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Guo C, Zhou B, Li C, Wang H, Zheng B, Ding H, Zhu Z, Peragine A, Cui Y, et al. Regulation of vegetative phase change by SWI2/SNF2 chromatin remodeling ATPase BRAHMA. Plant Physiol. 2016;172(4):2416–2428. doi: 10.1104/pp.16.01588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Lucas M, Pu L, Turco G, Gaudinier A, Morao AK, Harashima H, Kim D, Ron M, Sugimoto K, Roudier F, et al. Transcriptional regulation of Arabidopsis Polycomb repressive complex 2 coordinates cell-type proliferation and differentiation. Plant Cell. 2016;28(10):2616–2631. doi: 10.1105/tpc.15.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casamitjana-Martinez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol. 2003;13(16):1435–1441. doi: 10.1016/S0960-9822(03)00533-5. [DOI] [PubMed] [Google Scholar]
- 43.Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007;21(17):2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller K, Bouyer D, Schnittger A, Kermode AR. Evolutionarily conserved histone methylation dynamics during seed life-cycle transitions. PLoS One. 2012;7(12):e51532. doi: 10.1371/journal.pone.0051532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang C, Bratzel F, Hohmann N, Koch M, Turck F, Calonje M. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr Biol. 2013;23(14):1324–1329. doi: 10.1016/j.cub.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 46.Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131(21):5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- 47.Jiang D, Wang Y, Wang Y, He Y. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One. 2008;3(10):e3404. doi: 10.1371/journal.pone.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berger F, Chaudhury A. Parental memories shape seeds. Trends Plant Sci. 2009;14(10):550–556. doi: 10.1016/j.tplants.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Wang H, Liu C, Cheng J, Liu J, Zhang L, He C, Shen WH, Jin H, Xu L, Zhang Y. Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of Polycomb group proteins in association with distinct sets of Cis-regulatory elements. PLoS Genet. 2016;12(1):e1005771. doi: 10.1371/journal.pgen.1005771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carter B, Bishop B, Ho KK, Huang R, Jia W, Zhang H, Pascuzzi PE, Deal RB, Ogas J. The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in Arabidopsis. Plant Cell. 2018;30(6):1337–1352. doi: 10.1105/tpc.17.00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shu J, Chen C, Thapa RK, Bian S, Nguyen V, Yu K, Yuan ZC, Liu J, Kohalmi SE, Li C, et al. Genome-wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in Arabidopsis seedlings. Plant direct. 2019;3(1):e00100. doi: 10.1002/pld3.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mozgova I, Kohler C, Hennig L. Keeping the gate closed: functions of the polycomb repressive complex PRC2 in development. Plant J. 2015;83(1):121–132. doi: 10.1111/tpj.12828. [DOI] [PubMed] [Google Scholar]
- 53.Bratzel F, Lopez-Torrejon G, Koch M, Del Pozo JC, Calonje M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol. 2010;20(20):1853–1859. doi: 10.1016/j.cub.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 54.Molitor A, Shen WH. The polycomb complex PRC1: composition and function in plants. J Genet Genomics. 2013;40(5):231–238. doi: 10.1016/j.jgg.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 55.Schonrock N, Bouveret R, Leroy O, Borghi L, Kohler C, Gruissem W, Hennig L. Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 2006;20(12):1667–1678. doi: 10.1101/gad.377206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouveret R, Schonrock N, Gruissem W, Hennig L. Regulation of flowering time by Arabidopsis MSI1. Development. 2006;133(9):1693–1702. doi: 10.1242/dev.02340. [DOI] [PubMed] [Google Scholar]
- 57.Aichinger E, Villar CB, Di Mambro R, Sabatini S, Kohler C. The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell. 2011;23(3):1047–1060. doi: 10.1105/tpc.111.083352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 59.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8(5):532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 60.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 61.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12(12):799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 62.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 63.Saleh A, Al-Abdallat A, Ndamukong I, Alvarez-Venegas R, Avramova Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 2007;35(18):6290–6296. doi: 10.1093/nar/gkm464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pu L, Sung ZR. PcG and trxG in plants - friends or foes. Trends Genet. 2015;31(5):252–262. doi: 10.1016/j.tig.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 65.Xu F, Kuo T, Rosli Y, Liu MS, Wu L, Chen LO, Fletcher JC, Sung ZR, Pu L. Trithorax group proteins act together with a Polycomb group protein to maintain chromatin integrity for epigenetic silencing during seed germination in Arabidopsis. Mol Plant. 2018;11(5):659–677. doi: 10.1016/j.molp.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 66.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13(2):115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Madia VN, Benedetti R, Barreca ML, Ngo L, Pescatori L, Messore A, Pupo G, Saccoliti F, Valente S, Mai A, et al. Structure-activity relationships on Cinnamoyl derivatives as inhibitors of p300 histone Acetyltransferase. ChemMedChem. 2017;12(16):1359–1368. doi: 10.1002/cmdc.201700040. [DOI] [PubMed] [Google Scholar]
- 68.Hauser M-T, EJP B. Histochemical analysis of root meristem activity in Arabidopsis thaliana using a cyclin: GUS (β-glucuronidase) marker line. Plant Soil. 2000;226(1):1–10. doi: 10.1023/A:1026421417979. [DOI] [Google Scholar]
- 69.Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17(8):678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- 70.Lorrai R, Gandolfi F, Boccaccini A, Ruta V, Possenti M, Tramontano A, Costantino P, Lepore R, Vittorioso P. Genome-wide RNA-seq analysis indicates that the DAG1 transcription factor promotes hypocotyl elongation acting on ABA, ethylene and auxin signaling. Sci Rep. 2018;8(1):15895. doi: 10.1038/s41598-018-34256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Figure S1. Treatment with RDS 3434 reduces H3K27me3 protein level in the curly leaf-29 mutant.a, b Immunoblot of 5 days-old clf-29 seedlings directly grown with RDS 3434 (120 μM) or DMSO as control (a), and of DMSO- or RDS 3434-treated wild-type (Col) and clf-29 seedlings (b). Total proteins were probed with H3K27me3 specific antibodies, and H3 was used as loading control. Western blot (top) and densitometric analysis (bottom). In (b) is shown the relative H3K27me3 protein level (bottom left), and the ratio of DMSO- and 120 μM RDS 3434-treated clf-29/WT (bottom right). Results were obtained from two independent replicates with SD values. Significant differences were analyzed by t-test (*P ≤ 0.05).
Additional file 2. Figure S2. Treatment with RDS3434 of clf-29 mutant seeds affects seed germination. a Seed germination assays of clf-29 mutant seeds imbibed in the presence of RDS 3434 (120 μM) or DMSO as control. Germination rate was scored at 24, 36, 48 and 120 HAI (Hours After Imbibition). Data represent the mean of two independent biological replicates each performed in duplicate (25 seeds per replica). Significant differences were analyzed by t-test (*P ≤ 0.05, **P ≤ 0.01). b 5 days-old clf-29 mutant seedlings directly grown for 5 days in the presence of RDS 3434 (120 μM) or DMSO as control.
Additional file 3. Figure S3. Wild-type seedlings treated with 240 μM RDS 3434. 5 days-old wild-type (Ws-4) seedlings directly grown in the presence of of RDS 3434 (240 μM) or DMSO as control.
Data Availability Statement
The data sets supporting the results of this article are included within the article.