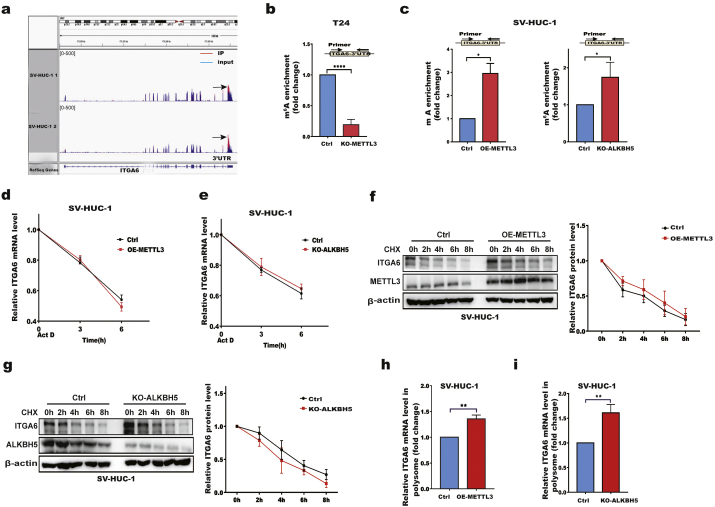
Fig. 3.
METTL3 and ALKBH5 regulate ITGA6 mRNA m6A modification and translation.
a m6A sites in the ITGA6 mRNA 3′UTR are shown from the of MeRIP-Seq data in SV-HUC-1 cells. b m6A enrichment in the ITGA6 mRNA 3′UTR was validated by MeRIP-qPCR in control and KO-METTL3 T24 cells. c m6A enrichment in the ITGA6 mRNA 3′UTR was validated by MeRIP-qPCR in control, OE-METTL3 and KO-ALKBH5 SV-HUC-1 cells. d, e ITGA6 mRNA stability in control, OE-METTL3 (d), and KO-ALKBH5 (e) SV-HUC-1 cells. RT-qPCR analysis of ITGA6 mRNA at the indicated time points after treatment with actinomycin D (Act D). f, g ITGA6 protein stability in control, OE-METTL3 (f), and KO-ALKBH5 (g) SV-HUC-1 cells. Western blotting of ITGA6 at the indicated time points after treatment with cycloheximide (CHX). h, i RT-qPCR analysis of the ITGA6 mRNA abundance in polysome fractions collected by sucrose gradient centrifugation in control, OE-METTL3 (h), and KO-ALKBH5 (i) SV-HUC-1 cells. All bar plot data are the means ± SEMs of three independent experiments except in (b, c), where the error bars denote the SDs of technical triplicates. *p < .05, **p < .01, ****p < .0001. (Student's t-test).