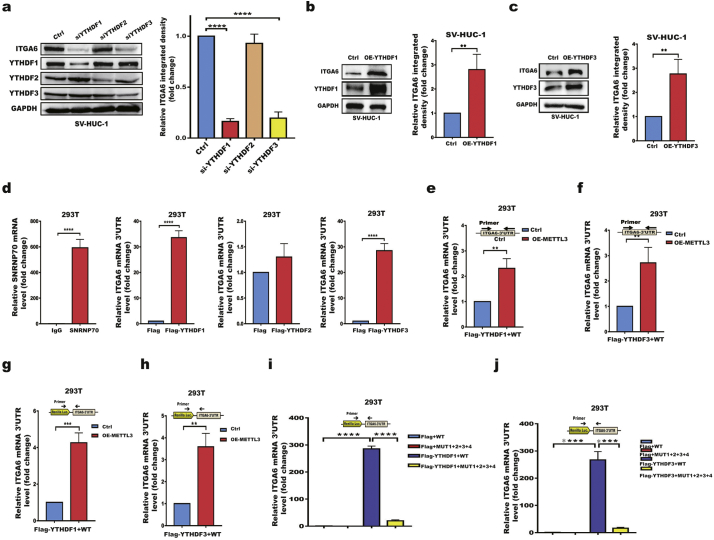
Fig. 5.
YTHDF1/YTHDF3 preferentially recognize m6A residues in the ITGA6 3′UTR and promote ITGA6 translation.
a Western blotting of ITGA6 expression in SV-HUC-1 cells treated with control or YTHDF1, YTHDF2, or YTHDF3 siRNAs. b, c Western blotting of ITGA6 in OE-YTHDF1 (b) or OE-YTHDF3 (c) SV-HUC-1 cells. d RIP analysis of the binding of the YTHDF1, YTHDF2, or YTHDF3 proteins to ITGA6 mRNA in 293T cells. FLAG-tagged YTHDF1, YTHDF2, YTHDF3 or control 2AB vectors were transfected into 293T cells. Lysates were immunoprecipitated with an anti-FLAG antibody. Enrichment of the ITGA6 mRNA 3′UTR with FLAG was measured by RT-qPCR and normalized to input. IgG, negative control, SNRNP70, positive control. e, f RIP analysis of the binding of the YTHDF1 (e) and YTHDF3 (f) proteins to ITGA6 mRNA in METTL3-overexpressing and control 293T cells g, h RIP analysis of the binding of the YTHDF1 (g) and YTHDF3 (h) proteins to exogenous ITGA6 mRNA 3′UTR. Both stable METTL3-overexpressing and control 293T cells were transfected with psiCHECK™-2-ITGA6 mRNA 3′UTR (WT) and FLAG-tagged YTHDF1, YTHDF3 or control 2AB vectors. Enrichment of the psiCHECK™-2-ITGA6 mRNA 3′UTR with FLAG was measured by RT-qPCR and normalized to input. The primer covers the junction between Renilla Luc and the ITGA6 3′UTR. i, j RIP analysis of the binding of the YTHDF1 (i) and YTHDF3 (j) proteins to exogenous ITGA6 mRNA 3′UTR containing wild-type m6A sites (WT) and 4 mutated m6A sites (MUT1 + 2 + 3 + 4). All bar plot data are the means ± SEMs of three independent experiments. **p < .01, ***p < .001, ****p < .0001. (Student's t-test, one-way ANOVA, Dunnett's test).