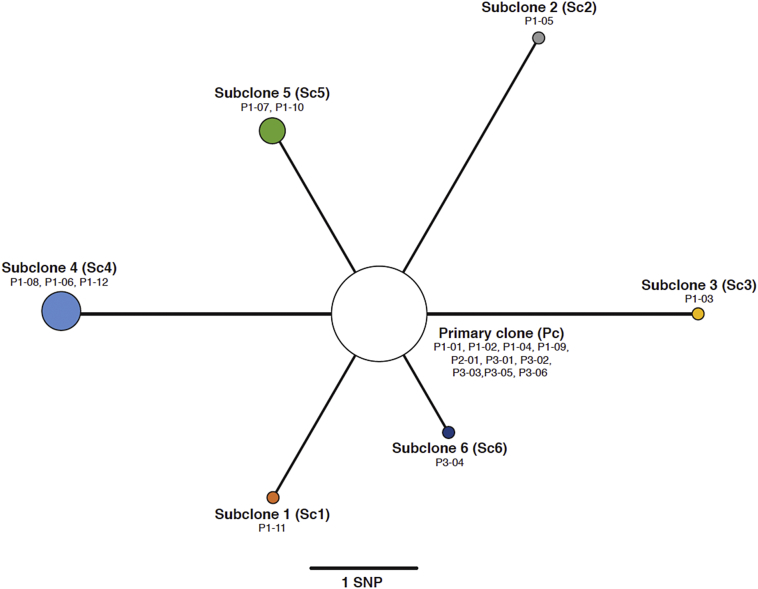
Fig. 1.
Maximum parsimony phylogeny drawn using high confidence fixed SNPs. Node sizes correspond to the number of samples representing each subclone population. We chose the most dominant clone (primary or subclone) from each sample. This represents an allele frequency was below 50% in some samples (P1-04, P1-07, P1-09, and P2-01), while still being the most abundant clonal population.