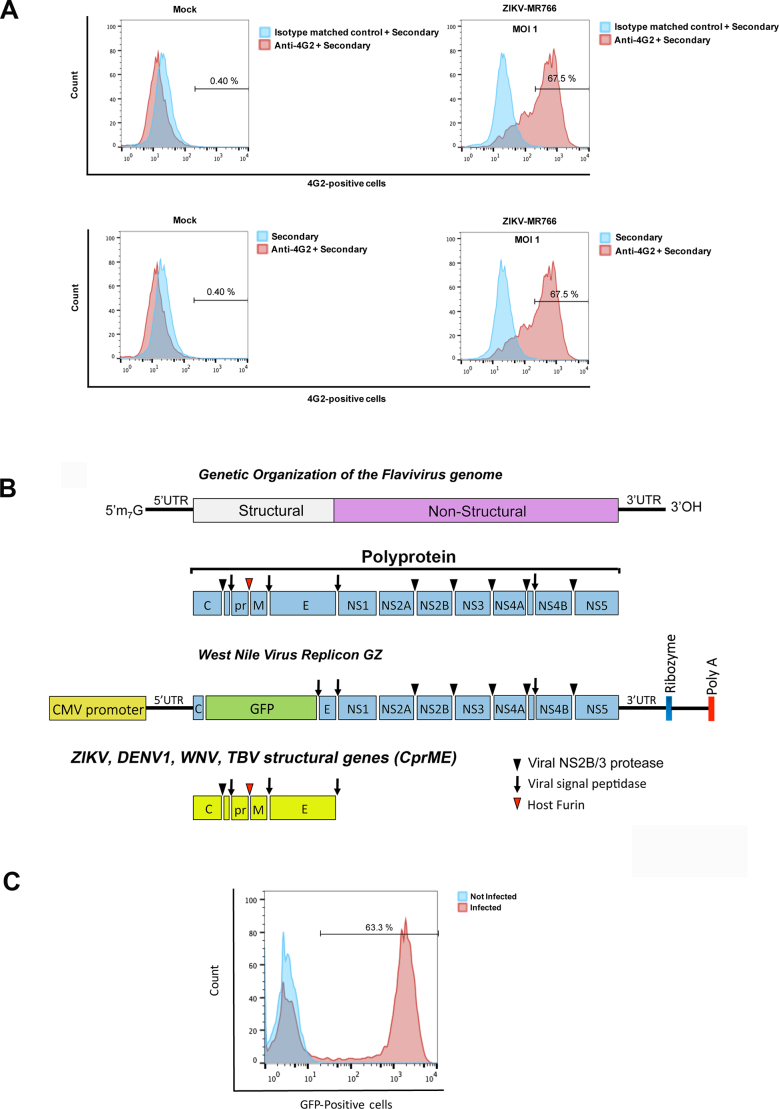
Fig. S1.
Detection of wild type ZIKV infection using the anti-envelope monoclonal antibody 4G2. (A) Vero cells were challenged by ZIKV strain MR766 (ZIKV-MR766) at an MOI of 1. Forty-eight hours post-challenge cells were fixed/permeabilized, and infection was determined by staining cells using anti- ZIKA envelope antibodies 4G2. The percentage of infected cells was measured by counting the number of 4G2-positive cells using a flow cytometer. As a control, fixed/permeabilized infected cells were also stained using an isotype matched control antibody. We performed the same experiments using fixed/permeabilized mock (non-infected) cells. (B) Construction of ZIKV, WNV, DENV1, TBV and JEV-reporter viral particles. The genetic organization of the Flavivirus genome is shown. The Flaviviruses genome is translated into a single polyprotein that is proteolytically processed into the illustrated structural and non-structural proteins. The West Nile Virus replicon GFP (GZ) is shown, which is the genome of West Nile virus where the GFP protein has been inserted replacing part of the capsid (C), promembrane (prM) and part of the envelope (E). The Zika Virus structural genes are shown. To create ZIKV-RVPs, human HEK293T cells were transfected with 5 μg of the West Nile virus replicon GZ together with 1 μg of the Zika Virus structural genes. Supernatants containing ZIKV- RVPs were collected 48 h post transfection, and ZIKV-RVPs were tittered in Vero cells using serial dilutions. (C) Forty-eight hours post-challenge infection was determined by measuring the percentage of GFP-positive cells using a flow cytometer.