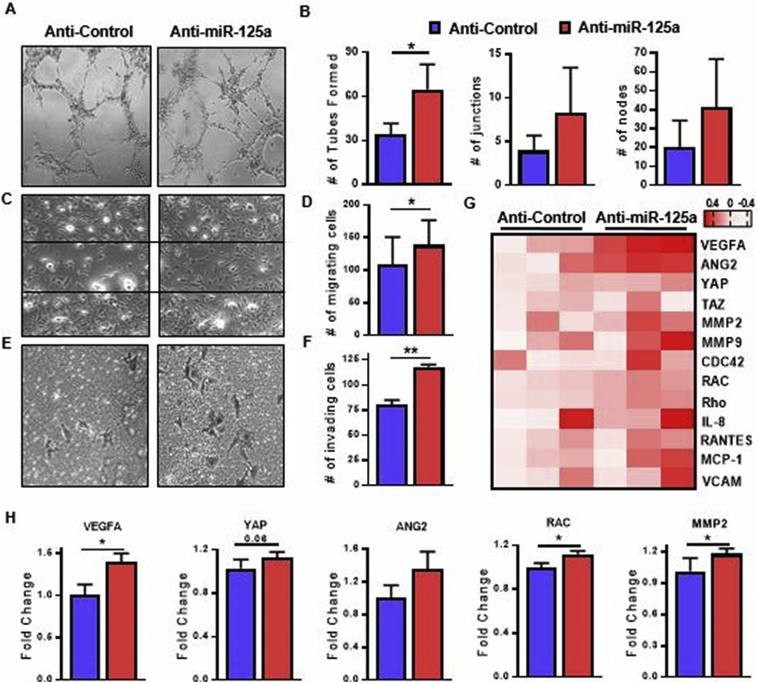
Fig. 2.
miR-125a promotes endothelial cell activation.
Representative images of (A) demonstrating HUVEC network formation in response to anti-miR-125 and scrambled controls (n = 3). (B) Bar graphs quantifying the number of tubes formed, branching points, junctions and nodes per high power field. (C) Representative photomicrographs demonstrating HUVEC wound repair assay in response to anti-miR-125a treatment compared to anti-control ECs (n = 3). (D) Bar graphs quantifying the number of migrating cells as counted in five random fields of vision. (E) Representative photomicrographs demonstrating HUVEC invasion in response to anti-miR-125 and scrambled controls (n = 3, original magnification 10×). (F) Bar graphs quantifying the number of migrating cells as counted in five random fields of vision. (G) Heatmap displaying the gene transcripts for the main molecular markers of endothelial cell activation as quantified by RT-PCR (n = 6), heatmap is presented as log2 values. (H) Gene sets of endothelial cell branching, migration and invasion compared in 3 anti-control/anti-miR-125a pairs. Relative gene expression measured by RT-PCR (n = 3). Data are represented as mean ± SEM. ** p < .01, * p < .05 significantly different compared to control. See also Fig. S1 and S2.