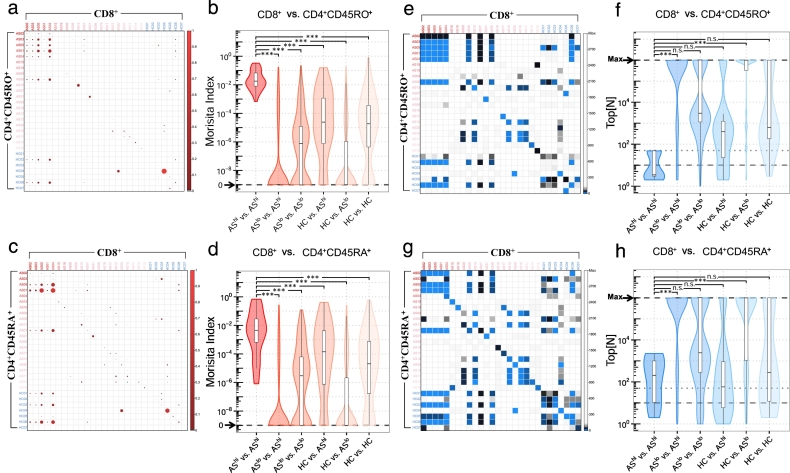
Fig. 3.
Similarity profiling reveals shared total TCRs (CDR3 nucleotides in combination with their corresponding V and J genes) between CD4+ and CD8+ in high disease activity AS patients. Morisita similarity indices for each pairwise combination of CD8+ and CD4 + CD45RO+ (a) or CD4 + CD45RA+ (c) samples plotted as heat maps. Red gradients denote MH values, which range from 0.0 (no shared clonotype between repertoires) to 1.0 (identical repertoires). Violin plots indicate the spread of Morisita similarity indices between CD8+ and CD4 + CD45RO+ (b) or CD4 + CD45RA+ (d). One-way ANOVA and Tukey's test were used to compare similarities across different groups. Top[N] index for each pairwise combination of CD8+ and CD4 + CD45RO+ (e) or CD4 + CD45RA+ (g) T cell samples plotted as heat maps. Blue gradients denote Top[N] values, which range from 1 (the very first shared clonotype from Top 1 clone between repertoires) to Max (no shared clonotype). Violin plots indicate the spread of Top[N] indices between CD8+ and CD4 + CD45RO+ (f) or CD4 + CD45RA+ (h) samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)