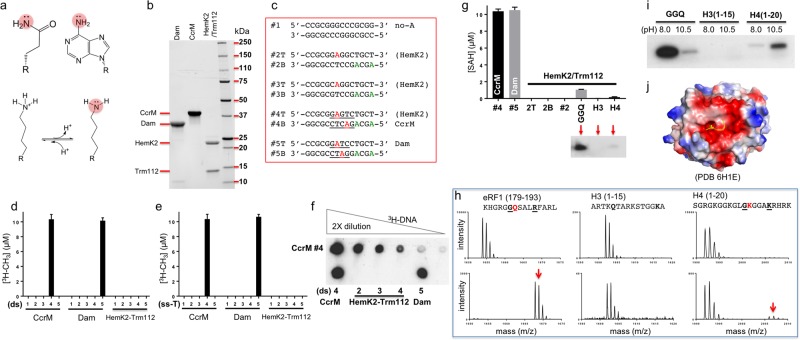
Fig. 1. Human HemK2-Trm112 complex methylates peptide substrates.
a Chemical structures of glutamine, adenine, and lysine. The target nitrogen atom is highlighted in red circle. b, c Recombinant proteins (panel b) and oligonucleotides (panel c) used in the study. d, e HemK2-Trm112 is not active on double-stranded DNA (panel d) or single-stranded DNA (panel e). f Fluorography of reaction products of 3H-DNA by indicated enzymes (bottom) and 2 × dilution of oligo #4 by CcrM (top). g HemK2-Trm112 is active on GGQ peptide substrates. The vertical axis indicates the reaction by-product (SAH) concentration, measured by the Promega MTase-GloTM assay. Autoradiography films of samples from reactions are shown in the panels below. h Representative spectra of MALDI-TOF before (top panels) and after reactions (bottom panels). i Effect of pH on the methylation of glutamine-containing and lysine-containing peptides shown by in-gel fluorography. j Surface representation of HemK2-Trm112 (PDB 6H1E) shows the substrate binding interface is enriched in negatively charged potential (red: negative; blue: positive, white: neutral). The H4 peptide is shown in yellow with the target lysine inserted into the active site where SAM binds