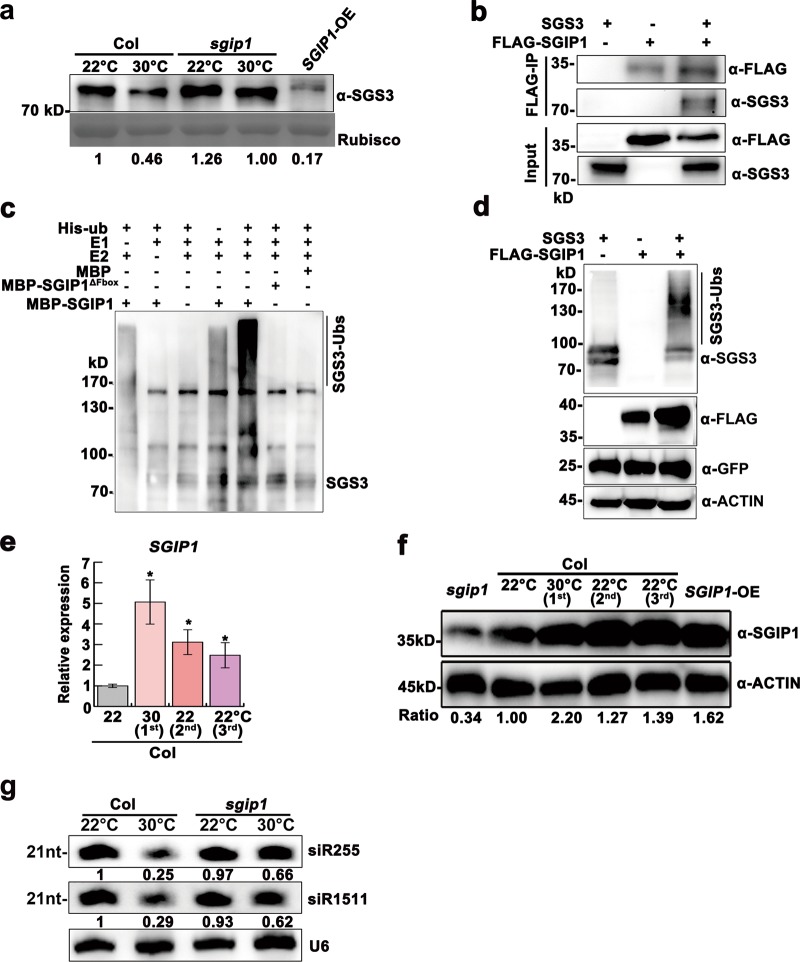
Fig. 2.
SGIP1 mediates thermomemory degradation of SGS3 and tasiRNA suppression. a Immunodetection of SGS3 abundance in 24-day-old plants. Coomassie blue staining of Rubisco served as a loading control. b Co-IP assays of SGIP1 with SGS3 in N. benthamiana leaves. Proteins were immunoprecipitated with an anti-FLAG antibody, and detected by western blot using the anti-SGS3 and anti-FLAG antibodies. c In vitro ubiquitination of SGS3 by SGIP1. Polyubiquitination of SGS3 by SGIP1 was detected by immunoblotting using the anti-SGS3 antibody. The mutated SGIP1 protein (SGIP1△Fbox) served as a negative control. d SGIP1 promotes SGS3 polyubiquitination and degradation in vivo. Co-expression of SGIP1 and SGS3 was performed in N. benthamiana leaves. Protein accumulation of SGS3 and FLAG-SGIP1 was immunodetected with the anti-SGS3 and anti-FLAG antibody, respectively. GFP was co-expressed as an internal control. e Expression levels of SGIP1 as normalized to the ACTIN2 signals. Error bars indicate the SD (n = 3). Asterisks indicate significant difference between lines and 22 °C-grown Col (Student’s t-test; *P < 0.05, **P < 0.01). f Immunodetection of SGIP1 levels with an anti-SGIP1 antibody. ACTIN served as a loading control (d, f). g Knockdown of SGIP1 maintained the production of siR255 and siR1511 at 30 °C. U6 was used as a loading control. The intensity of the blots was quantified (a, f, g)