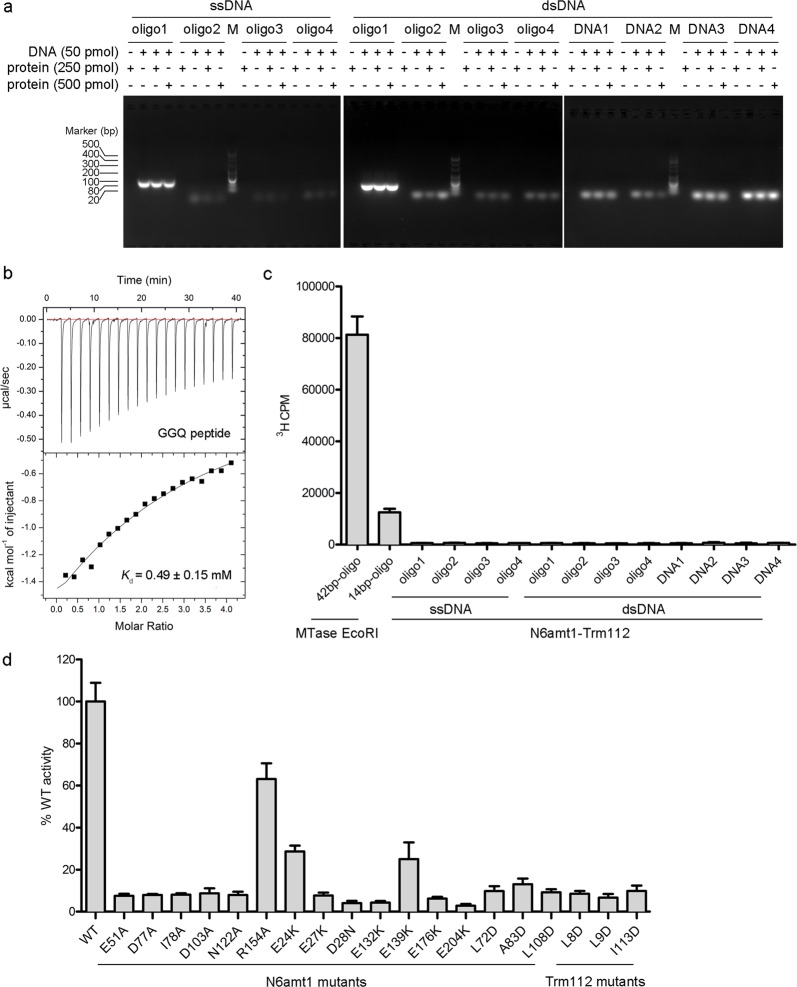
Fig. 3. Biochemical studies of the N6amt1–Trm112 complex.
a EMSA analysis of N6amt1–Trm112 towards different ssDNA and dsDNA. Oligos1–4 are the four DNA oligos used by Xiao et al. in the examination of the DNA 6mA modification activity of N6amt110. The other four oligos are as follows: DNA1, CTTTTTAGAAGCACA; DNA2, AATGAGAAGTTTATC; DNA3, AAAATCGAGGTTCCC; DNA4, ACTGACCATGGATCG. b ITC analysis of the binding affinity of N6amt1-Trm112 towards the 15-residue GGQ peptide of eRF1. Prior to titration, the protein complex was incubated with SAH. The dissociation constant (Kd) is indicated. c Enzymatic activity assay of N6amt1–Trm112 towards dsDNAs and ssDNAs with different lengths and sequences. MTase EcoRI is used as a positive control. All the experiments were performed in triplicates and the error bars represent the standard error of the mean. d Effects of mutations of the key residues at the active site of N6amt1 and the N6amt1–Trm112 interface on the enzymatic activity of N6amt1-Trm112 for the methylation of human eRF1. The average of 3H incorporation measurement of the wild-type protein complex (about 2300 c.p.m.) is defined as “100% WT activity”, and the MTase activities of the N6amt1–Trm112 mutants are shown relative to 100% WT activity. All the experiments were performed in triplicates and the error bars represent the standard error of the mean