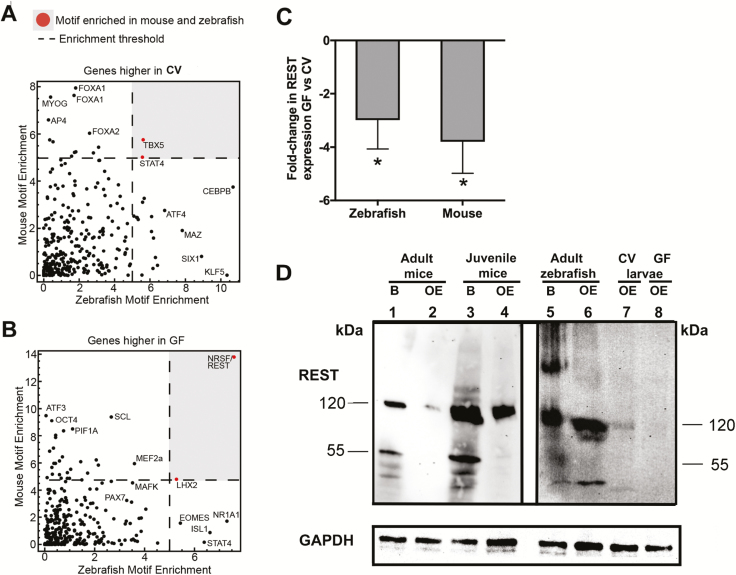
Figure 3.
NRSF/REST-binding sites are commonly enriched at genes that are downregulated in olfactory tissue in response to microbiota. (A) TF motif enrichment (−log10P-value) using the 1000 bp upstream of the TSS of genes that are significantly more highly expressed in CV conditions in zebrafish and mouse olfactory tissue. A dashed line represents the threshold for motif enrichment. TBX5 and STAT4 motifs that are enriched in both zebrafish and mouse are marked in red and within the gray area. (B) Same as (A) for genes that were significantly higher in GF conditions. NRSF/REST and LHX2 motifs that are enriched in both zebrafish and mouse are marked in red and within the gray area (P < 0.05). (C) Gene expression levels of REST in GF zebrafish and mouse compared to CV animals (N = 4). Asterisk indicates statistically significant differences when P < 0.05. (D) Protein expression of REST in brain and olfactory organ of adult mice (lanes 1–2), juvenile mice (lanes 3–4), adult zebrafish brain and olfactory organ (lanes 5–6), and olfactory organs of CV and GF zebrafish larvae (lanes 7–8). Blots probed with an isotype control instead of anti-REST antibody produced no bands. GAPDH was used as a loading control.