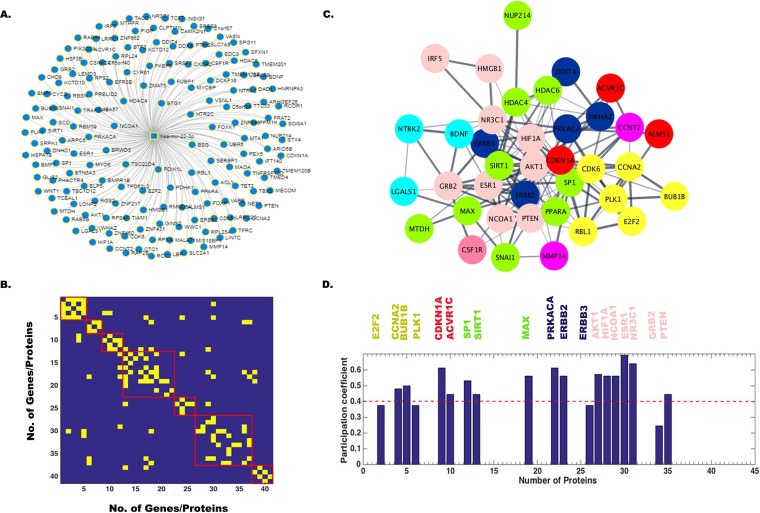
FIG 3.
Network analysis showing miR-22-3p target genes, modularity/community detection in gene networks, and the identification of hub genes based on participation coefficients. (A) A comprehensive human miRNA-gene network constructed in miRnet.ca database. In String database, 40 genes were found to be connected, and based on their interaction, an adjacency matrix Aij was formed, which was used for network analysis. This adjacency matrix Aij comprises 40 target genes (nodes) and binarized edges based on their functional annotations (B). Modularity score Q was computed using the Brain Connectivity Toolbox (BCT) in Matlab, and genes were partitioned into seven identified communities based on similarity of connection patterns for each pair of vertices/nodes (C). Based on the node degree kv of node v, and kvs being the number of edges from the vth node to nodes within module s, we have estimated each vertex’s participation index P and quantified the presence of provincial and connector hub genes (D). The participation index P shows that there are provincial and connector hub genes belonging to seven communities.