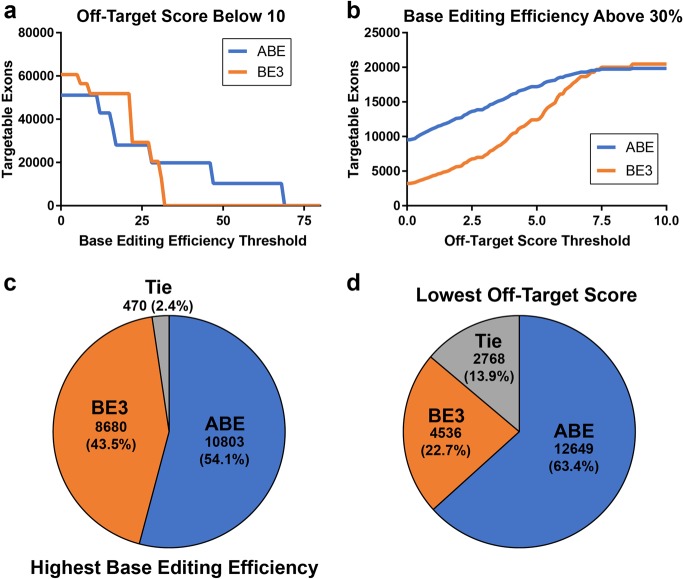
Fig. 8. ABE is predicted to have improved on-target and off-target editing efficiencies compared to BE3 when targeting splice acceptor sites.
a Genome-wide computational analysis of the number of inner exons that can be targeted by ABE and BE3 with predicted editing efficiency of the target base at or above the value on the x-axis. Only sgRNAs with an off-target score below ten were considered. b Estimation of the number of inner exons that can be targeted by ABE and BE3 using sgRNAs with off-target scores at or below the value on the x-axis. Only sgRNAs with an on-target base editing efficiency above 30% were considered. c A total of 19,953 inner exons in the human genome can be targeted by both ABE and BE3. The sgRNAs needed to induce skipping with ABE have higher predicted base editing efficiency for targeting 54.1% of the exons, d and lower predicted off-target score for targeting 63.4% of the exons