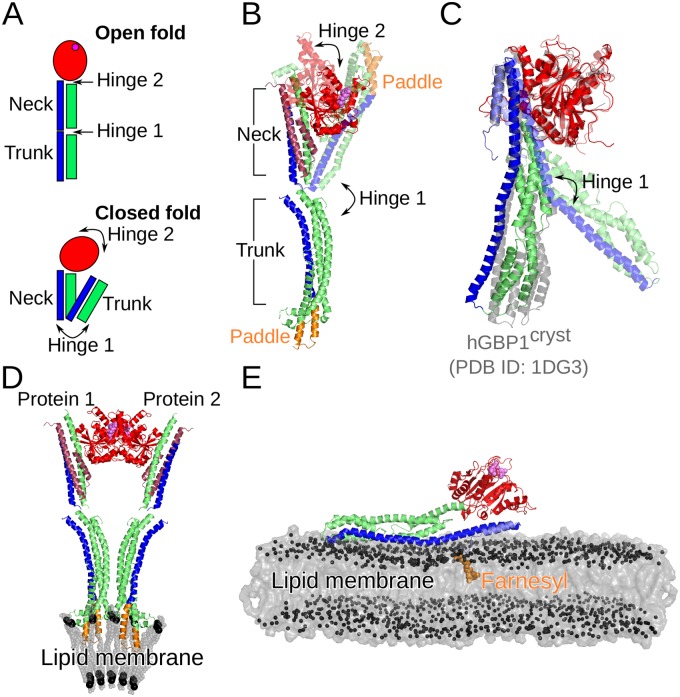
Fig 8. Conformational transitions of hGBP1 and a bacterial dynamin-like protein (BDLP) of Nostoc punctiforme.
(A) Schematic providing nomenclature for the open and closed BDLP conformations with large-scale transitions between them mediated by hinges 1 and 2 at the trunk/neck and neck/G-domain interfaces. As for hGBP1, the GTPase domain is colored in red with the position of the nucleotide in the open conformation indicated by a magenta sphere, while the 4-helix bundle composing the neck and trunk is colored in green and blue. (B) The closed conformation of BDLP was solved by X-ray crystallography (PDB 2J69, shown in transparent cartoon), while the open conformation was determined by cryo-electron microscopy (PDB 2W6D, shown in opaque cartoon). The two helices preceding the G domain are ruby-colored. (C) From our MD simulations of hGBP1 we observed a kinking of the M and E domain resembling the initial stages of the closed ↔ open transition of BDLP around hinge1, as the overlay of the structures representing this motion (i.e., FEM1 and PC1min in Fig 4) shows. For comparison the crystal structure of hGBP1 (PDB 1DG3) is also shown (in gray). (D) Membrane binding of BDLP occurs following nucleotide binding via the paddle domain (shown in orange) in the open conformation and aggregated BDLP state. Aggregation involves association of the G domains. (E) Membrane binding of hGBP1 also requires nucleotide binding and is facilitated via farnesylation (shown in orange) at the C-terminus acting as lipid anchor.