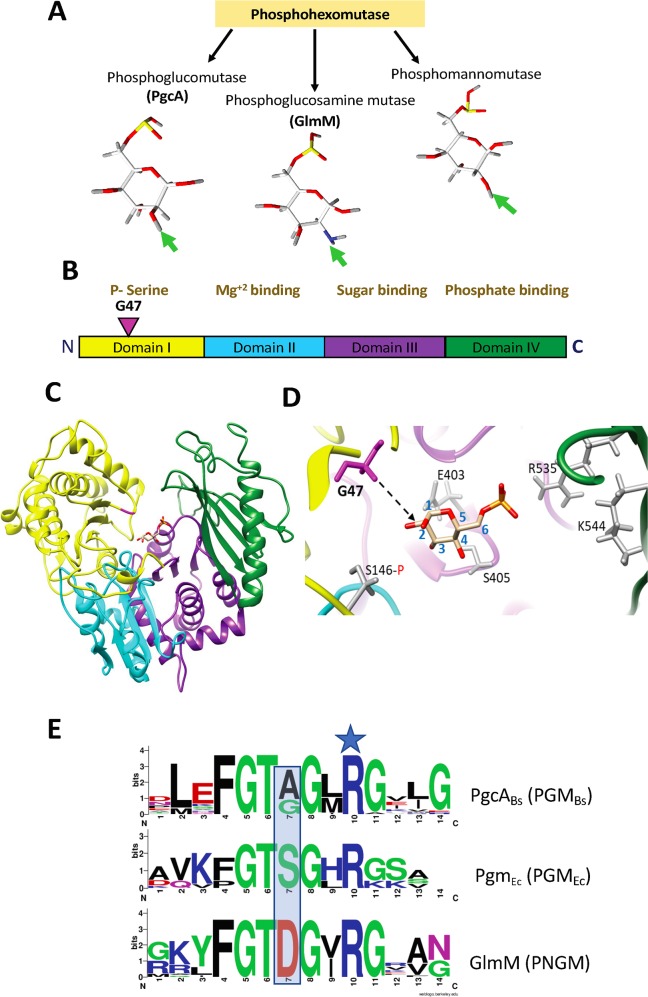
Fig 6. Identification of a mutation affecting substrate-selectivity in PGM/PNGM family enzymes.
(A) Schematic depicting enzymes from the phosphohexomutase family and their 6-phosphohexose sugar substrates, with the variable substituents at the C2 position highlighted by an arrow. (B) domain structure and conserved functions within PGM enzymes, as detailed in [22]. N and C indicate the N-terminus and C-terminus of the enzyme, respectively. The pink inverted triangle indicates residue G47 in domain I. (C) B. subtilis PGM (PgcA) modelled with I-TASSER using P. aeruginosa bifunctional PGM/PMM (PDB#1P5G; which contains bound glucose 6-phosphate) as a scaffold (25% identical to B. subtilis PgcA) illustrating the position of the bound glucose 6-phosphate in the substrate-binding cleft. (D) Enlarged view of the substrate-binding site of modeled B. subtilis PgcA. Residue G47 is illustrated in pink with a black broken arrow indicating the relationship of the protein backbone in this region to the carbon-2 position of bound glucose 6-phosphate. The grey residues include functionally important residues of the active site including the serine (S146) that is involved in phosphoryl transfer (also located in domain I), as well as previously described sugar phosphate binding residues (E403, S405, R535, K544; B. subtilis numbering) of domain IV. As per the homology modelling, E403 and S405 would interact with the sugar backbone, whereas R535 and K544 form salt bridges with the bound glucose 6-phosphate. (E) Sequence Logo representation of conserved amino acids in the loop region adjacent to an invariant active site Arg residue (blue star) in three groups of PGM/PNGM enzymes including 24 bacterial PGM enzymes related to B. subtilis PgcA (designated as the PGMBs group) and defined by a conserved motif with an Ala/Gly at the position corresponding to PgcA Gly47 (residue 7 in the top motif), 8 proteobacterial PGM enzymes including E. coli Pgm (designated as the PGMEc group) defined by a Ser residue at this position (middle motif), and 20 bacterial PNGM (GlmM) enzymes with an Asp at this position. Sequence Logos were derived from the aligned sequences in S4 and S5 Figs.