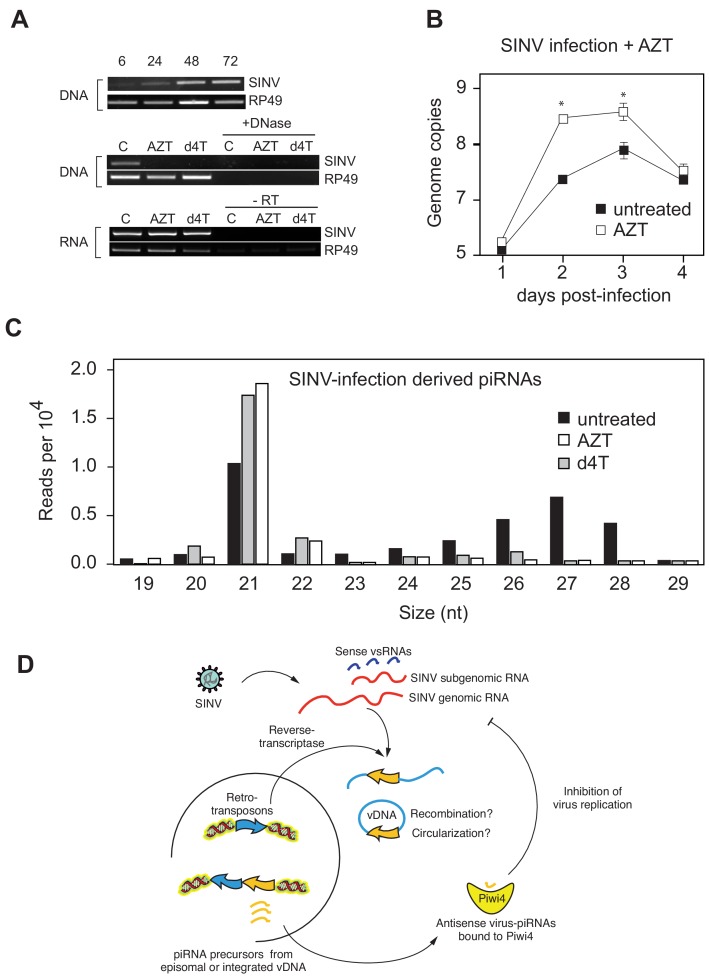
Figure 4. Production of v-piRNAs is blocked by reverse-transcriptase inhibitors.
(A) Multi step growth curve of SINV RNA in Aag2 cells (MOI 0.1) with or without AZT treatment measured by RT-qPCR. RP49 was used as a normalization control. The error bars represent standard deviation of four biological replicates. Significant changes over untreated control are marked with asterisks (p<0.05, Mann-Whitney U test). B Size distribution plot of small RNAs mapping to the SINV genome from untreated infected Aag2 cells or cells treated with the RT-inhibitors AZT or d4T. Read counts per hundred were normalized to SINV genome copy numbers. (C) Schematic representation of the v-piRNA production. vDNA localized in the nucleus where it is transcribed to produce primary piRNA transcripts that are processed by and bound to Piwi4, and transported to the cytoplasm where they participate in the targeting of viral RNA.